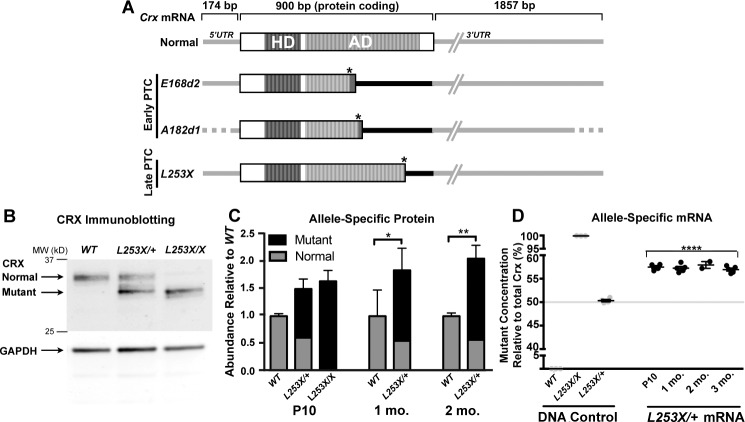
Figure 1.
Class III Crx mutations introduce a PTC, resulting in accumulation of truncated CRX proteins and mutant mRNA with variable extended 3′UTR. (A) Schematic representation of normal and mutant Crx mRNA species, showing untranslated (UTR, gray/black lines) and protein-coding (open/striped boxes) with their relative sizes (bp), based on mouse (Normal, E168d213 and L253X) and cat (A182d1)15 models. The DNA-binding homeodomain (HD, dark strips) and the activation domain (AD, light strips) sequences12 are indicated. All three mutant mRNAs encode the complete HD, but ADs are truncated at different positions due to their PTC (indicated as the right-hand end of the protein coding box) induced by the mutation (marked by an asterisk). These PTCs also expand the 3′UTR to include different lengths of the original coding region (indicated by black line) in front of the normal 3′UTR (gray line). The position of the PTC relative to the normal stop codon determines the length of expanded 3′UTR: Early PTC mutants, E168d2 and A182d1 (Rdy cat mutation) produce a longer 3′UTR than the late PTC mutant L253X. Since 5′UTR and 3′UTR sequences of feline Crx mRNA are not available, these regions of the A182d1 transcript are indicated by dotted lines. (B) Immunoblot of CRX in the retinas of WT, L253X/+, and L253X/X mutants at 1-month old (mo) with GAPDH as a protein loading control. Black arrows indicate the running position for the normal and mutant (truncated) forms of CRX. (C) Quantification of relative amounts of normal and mutant CRX proteins in the indicated retinas at three ages (mean ± SEM, n ≥ 3; * P < 0.05, ** P < 0.01; 2-Way ANOVA with Tukey's multiple comparisons test). (D) ddPCR quantification of mutant (L253X) and normal Crx mRNA as percentage of total Crx. The specificity of the two allele-specific ddPCR assays was established using triplicate tail DNA samples (gray circles) from the indicated genotypes (DNA control). L253X/+ mRNA results are presented as percent of total Crx mRNA (mutant plus normal transcripts; mean ± SEM, n ≥ 3; **** P < 0.0001; unpaired t-test for mutant mRNA level relative to L253X/+ DNA control; n ≥ 3).