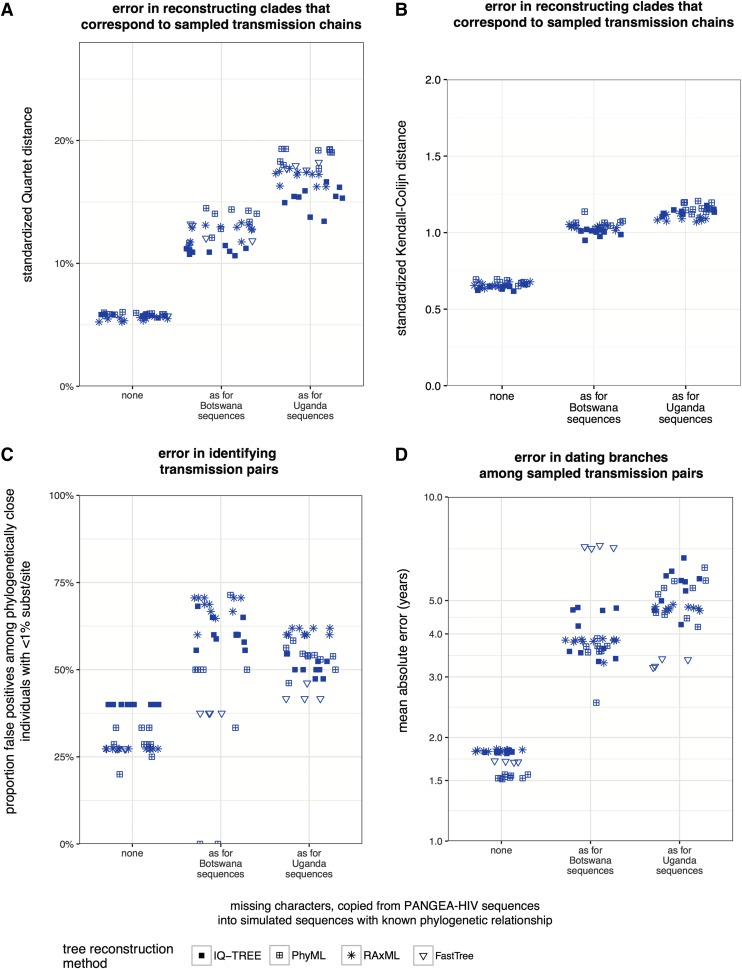
FIG. 3.
Impact of missing characters in PANGEA-HIV sequences on phylogeny reconstruction when sequences are sparsely sampled. Three sequence data sets of 1,600 taxa of concatenated HIV-1 gag, pol, env genes were simulated. For each data set, missing characters in real PANGEA-HIV sequences from specific sampling locations (see x-axis) were copied into simulated sequences (data sets D1–D3, see Supplementary Table S1). Phylogenies were reconstructed in replicate with several tree reconstruction algorithms and compared to the true phylogeny. (A) Quartet distance between reconstructed and true subtrees that correspond to sampled transmission chains in the simulations. (B) Kendall-Colijn distance between reconstructed and true subtrees that correspond to sampled transmission chains in the simulations. (C) Proportion of false-positive transmission pairs among pairs of individuals that diverged less than 1% substitution/site in reconstructed phylogenies. (D) Mean absolute error (years) in estimated divergence times between sequences from sampled transmission pairs. Across all error measures, reconstructed phylogenies were considerably less accurate when sequences were sparsely sampled and contained missing characters as seen among PANGEA-HIV sequences from Botswana or Uganda, compared to gag+pol+env sequences without missing characters.