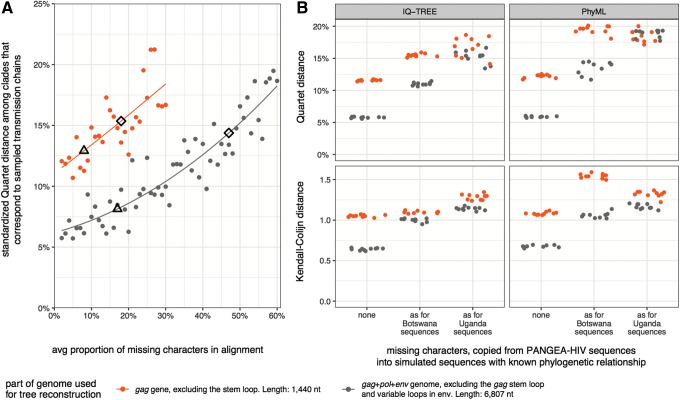
FIG. 5.
Alignment trimming to reduce tree reconstruction artifacts. (A) Sixty alignments of 1,600 gag+pol+env sequences (6,807 nt) with increasing proportions of missing characters were simulated. Missing site patterns were copied at random from PANGEA-HIV sequences (data sets D1-Mxx, see Supplementary Table S1). Thirty alignments were trimmed to the gag gene. One phylogeny per alignment was reconstructed with RAxML. We compared Quartet distances of trees reconstructed from patchy gag+pol+env sequences (gray) to those of patchy gag sequences (orange). It is possible to reconstruct more accurate phylogenies from shorter gag sequences, but only when the trimmed alignment harbors substantially fewer missing characters than the longer original alignment and sequence sampling coverage is low (6%). The proportion of missing characters in gag and gag+pol+env sequences among PANGEA-HIV sequences from Botswana and Uganda is indicated with triangles and diamonds. (B) The three sequence data sets of 1,600 gappy gag+pol+env sequences of Figure 2 were trimmed to the gag gene. Ten phylogenies were reconstructed with IQ-TREE, PhyML, and RAxML per alignment, and results are shown for IQ-TREE and PhyML. Tree reconstructions from gag genes that harbored missing characters as seen in PANGEA-HIV sequences from Botswana or Uganda were not more accurate than those from patchy gag+pol+env sequences, regardless of distance measure and tree reconstruction method. The differences in missing character patterns between the trimmed and original alignments were not large enough to result in more accurate tree reconstructions with the trimmed alignment.