Abstract
Consumption of ergot alkaloid-containing tall fescue grass impairs several metabolic, vascular, growth, and reproductive processes in cattle, collectively producing a clinical condition known as “fescue toxicosis.” Despite the apparent association between pituitary function and these physiological parameters, including depressed serum prolactin; no reports describe the effect of fescue toxicosis on pituitary genomic expression profiles. To identify candidate regulatory mechanisms, we compared the global and selected targeted mRNA expression patterns of pituitaries collected from beef steers that had been randomly assigned to undergo summer-long grazing (89 to 105 d) of a high-toxic endophyte-infected tall fescue pasture (HE; 0.746 μg/g ergot alkaloids; 5.7 ha; n = 10; BW = 267 ± 14.5 kg) or a low-toxic endophyte tall fescue–mixed pasture (LE; 0.023 μg/g ergot alkaloids; 5.7 ha; n = 9; BW = 266 ± 10.9 kg). As previously reported, in the HE steers, serum prolactin and body weights decreased and a potential for hepatic gluconeogenesis from amino acid-derived carbons increased. In this manuscript, we report that the pituitaries of HE steers had 542 differentially expressed genes (P < 0.001, false discovery rate ≤ 4.8%), and the pattern of altered gene expression was dependent (P < 0.001) on treatment. Integrated Pathway Analysis revealed that canonical pathways central to prolactin production, secretion, or signaling were affected, in addition to those related to corticotropin-releasing hormone signaling, melanocyte development, and pigmentation signaling. Targeted RT-PCR analysis corroborated these findings, including decreased (P < 0.05) expression of DRD2, PRL, POU1F1, GAL, and VIP and that of POMC and PCSK1, respectively. Canonical pathway analysis identified HE-dependent alteration in signaling of additional pituitary-derived hormones, including growth hormone and GnRH. We conclude that consumption of endophyte-infected tall fescue alters the pituitary transcriptome profiles of steers in a manner consistent with their negatively affected physiological parameters.
Introduction
Epichloe coenophialum is an endophytic fungus that infects most tall fescue (Lolium arundinaceum) pastures commonly used in animal grazing systems in the eastern half of the United States [1]. The interaction between N. coenophialum and tall fescue produces ergot alkaloids [2]. Consumption of ergot alkaloid-containing tall fescue impairs several metabolic, vascular, growth, and reproductive processes in cattle, collectively producing a clinical condition known as “fescue toxicosis” [3].
The anterior pituitary gland secretes hormones that affect control over several physiological processes altered by consumption of ergot alkaloid-containing forages, including hormones for metabolism (TSH), growth (GH), reproduction (LH, FSH), stress responses (ACTH), and lactation (prolactin) [4]. Despite these known relationships, we are unaware of reports that describe the effect of fescue toxicosis on pituitary genomic expression profiles.
The goal of the current research was to determine whether gene expression profiles differed between whole pituitaries of growing beef steers grazing pastures containing high (HE) or low (LE) amounts of toxic endophyte-infected tall fescue. We used transcriptome and targeted gene expression analyses to identify specific candidate molecules and signaling pathways responsible for the altered physiology of steers consuming HE forages. The global hypothesis tested was that consumption of endophyte-infected tall fescue would alter pituitary transcriptome profiles and that at least the pituitary genes responsible for the production and secretion of prolactin would be down-regulated and those for POMC/ACTH would be up-regulated.
Materials and methods
Animal model
All procedures involving animals were approved by the University of Kentucky Institutional Animal Care and Use Committee. The animal management regimen and model for steers that yielded the pituitary tissue of the present experiment have been reported. As described in detail previously [5–7], 19 beef steers (predominately Angus) were denied access to feed and water for 14 h, weighed, and subdivided into 2 groups based on BW. The steers were randomly allotted (d0) within BW group to graze either a low-toxic endophyte tall fescue-mixed pasture (LE; 5.7 ha; 0.023 μg ergot alkaloids/g; n = 9; BW = 267 ± 14.5 kg) or a high-toxic endophyte-infected tall fescue pasture (HE; 5.7 ha; 0.746 μg ergot alkaloids/g; n = 10; BW = 266 ± 10.9 kg) for an 89-d grazing period. Analysis of ergot alkaloid levels between the two pastures revealed that the HE steers were exposed to 25 and 21 times more ergovaline/ergovalinine and lysergic acid/isolysergic acid, respectively, than were the LE steers [5]. After the common 89-d grazing period on pastures, steers were slaughtered in the University of Kentucky Meat Laboratory (Lexington, KY) over a 17-day period. Throughout the slaughter period, steers continued to graze their respective treatment pastures. Details of the slaughter period and process have been reported [5].
Sample collection and RNA preparation
Steers were stunned by captive bolt pistol and exsanguinated. Within 10 to 12 minutes of death, the whole pituitary was collected from each animal, placed in a foil pack, flash-frozen in liquid nitrogen, and stored at -80°C. Three pituitary glands (1 LE, 2 HE) were not used in the microarray analysis because of tissue damage incurred during the collection process. As a result, eight pituitaries (n = 8) for both LE and HE treatment groups were subjected to RNA analyses.
Total RNA was extracted from the whole frozen pituitary tissue using TRIzol Reagent (Invitrogen Corporation, Carlsbad, CA) following the manufacturer’s instructions. The RNA concentrations were determined using a NanoDrop ND-1000 Spectrophotometer (NanoDrop Technologies, Wilmington, DE), which revealed that all samples had an average concentration of 678 ng/μl and were of high purity with 260:280 nm absorbance ratios ranging from 1.71 to 1.91 and 260:230 nm absorbance ratios ranging from 2.08 to 2.55. The integrity of total RNA was examined by gel electrophoresis using an Agilent 2100 Bioanalyzer System (Agilent Technologies, Santa Clara, CA) at the University of Kentucky Microarray Core Facility. All RNA samples had 28S:18S rRNA absorbance ratios greater than 1.7 and RNA integrity numbers greater than 8.7.
Microarray analysis
The custom WT Btau 4.0 Array (version 1) GeneChip (Affymetrix, Inc., Santa Clara, CA) was used [8] to investigate the effect of HE vs. LE consumption on bovine pituitary gene expression profiles. Microarray analysis was conducted according to the manufacturer’s standard protocol at the University of Kentucky Microarray Core Facility. Briefly, 3 μg of RNA for each sample was first reverse-transcribed (RT) to cDNA and then from cDNA (double-stranded) to complementary RNA (cRNA; single-stranded), which was then labeled with biotin. The biotinylated cRNAs were further fragmented and used as probes to hybridize the gene chips in the GeneChip Hybridization Oven 640 (Affymetrix), using 1 chip per RNA sample. After hybridization, the chips were washed and stained on a GeneChip Fluidics Station 450 (Affymetrix). The reaction image and signals were read with a GeneChip Scanner (GCS 3000, 7G; Affymetrix), and data were collected using the GeneChip Operating Software (GCOS, version 1.2; Affymetrix). The raw expression intensity values from the GCOS (i.e., 16 *.cel files from the raw methylation measurements) were imported into Partek Genomics Suite software (PGS, version 6.6; Partek Inc., St. Louis, MO). For GeneChip background correction, the algorithm of Robust Multichip Averaging adjusted with probe length and GC oligo contents was implemented [9, 10]. The background-corrected data were further converted into expression values using quantile normalization across all the chips and median polish summarization of multiple probes for each probe set.
All the GeneChip transcripts were annotated using the NetAffx annotation database for Gene Expression on Bovine GeneChip Array ST 1.1, provided by the manufacturer (http://www.affymetrix.com/analysis/index.affx, last accessed in March 2016, annotation file last updated in April 2014). Quality control of the microarray hybridization and data presentation was performed by MA plot on all the gene expression values and by box plot on the control probe sets on the Affymetrix chips. Pearson (Linear) Correlation generated the similarity matrix (last accessed in March 2016, Partek Genomics Suite 6.6 6.15.0422). The average correlation between any pair of the 16 GeneChips was 0.98, and all GeneChips were further analyzed. Principal component analysis (PCA) was performed to elucidate the quality of the microarray hybridization and visualize the general data variation among the chips (Partek, 2015). To assess treatment effects (HE vs. LE) on the relative expression of the pituitary gene transcripts, qualified microarray data were subjected to one-way ANOVA using the same PGS software. To achieve a higher degree of confidence (i.e., a more conservative approach), transcripts showing treatment effects at the significance level of P < 0.001 (false discovery rate of ≤ 4.8%) were defined as differentially expressed. These differentially expressed genes/gene transcripts (DEGs) were subjected to hierarchical clustering analysis using PGS software and to canonical, functional, and network pathway analyses using the Core Analysis program of Ingenuity Pathway Analysis online software (IPA, Build version 430059M, Content version 31813283; http://www.ingenuity.com [accessed in December, 2016]; Ingenuity Systems, Inc., Redwood City, CA).
All the microarray *.cel files collected by GCOS plus the GC Robust Multichip Averaging-corrected data processed by PGS software of this manuscript have been deposited in the National Center for Biotechnology Information’s Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) [released October 23, 2014]), are minimum information about a microarray experiment (MIAME) compliant [11], and are accessible through GEO series accession number GSE62570.
Real-time RT-PCR analysis
Primer sets for genes selected for real-time reverse transcription (RT) PCR analysis (S1 Table) were designed using the NCBI Pick Primers online program against RefSeq sequences (accessed January to June 2016). Real-time RT-PCR was performed using an Eppendorf Mastercycler ep realplex2 system (Eppendorf, Hamburg, Germany) with iQ SYBR Green Supermix (Bio-RAD, Hercules, CA), as described [12]. Briefly, cDNA was synthesized using the SuperScript III 1st Strand Synthesis System (Invitrogen), with 1 μg of RNA used for each reverse transcription reaction. Real-time RT-PCR was performed with a total volume of 25 μL per reaction, with each reaction containing 5 μL of cDNA, 1 μL of a 10 μM stock of each primer (forward and reverse), 12.5 μL of 2× SYBR Green PCR Master Mix, and 5.5 μL of nuclease-free water. Gene expression was analyzed by the 2−ΔΔCT method [13].
The resulting real-time RT-PCR products were purified using a PureLink Quick Gel Extraction Kit (Invitrogen) and sequenced at Eurofins Scientific (Eurofins, Louisville, KY). Sequences were compared with the corresponding RefSeq mRNA sequences used as the templates for primer set design. The sequences of the primers and the resulting sequence-validated real-time RT-PCR reaction amplicons for selected DEGs and the endogenous control genes ACTB, PPIA, and UBC are presented in S1 Table and S1 Fig, respectively. Primers for ACTB were from Lisowski et al. [14], and primers for s-PRLR and l-PRLR were from Thompson et al. [15]. All sequenced amplicons had at least 98% identity with their template sequences. The raw CT values of ACTB, PPIA, and UBC in pituitary tissue of HE and LE steers did not differ (P = 0.57, 0.42, 0.82; respectively). Accordingly, the geometric mean expression of ACTB, PPIA, and UBC was used to normalize the relative quantities of the selected DEGs mRNA expression, and all RT-PCR reactions were conducted in triplicate.
Selected miRNA-target gene interactions
To identify (predict) microRNAs (miRNAs) that might regulate [15, 16] prolactin or POMC/ACTH production, microarray-identified differentially expressed miRNAs (DEMs) were uploaded into TargetScan online software (Release 7.1, http://www.targetscan.org/), and the species-specific “Cow” filter applied. The resulting miRNA candidates were ranked based on cumulative weighted context++ scores [16] and then reduced to only those predicted to bind mRNA of genes involved in prolactin or POMC/ACTH production or to bind to mRNA coding known transcription factors of prolactin and POMC/ACTH pathway genes.
Statistical analyses
To test for HE vs. LE treatment effects on the relative expression of the pituitary gene transcripts, microarray hybridization data were subjected to one-way ANOVA using the PGS software as described in the “Microarray Analysis” section above. To determine the effect of treatment, the relative expression levels of selected DEGs analyzed by real-time RT-PCR were subjected to one-way ANOVA using the GLM procedure of the SAS statistical software package (version 9.4; SAS Inst., Inc., Cary, NC), with the endophyte level as the fixed effect. For these data, significance was declared when P ≤ 0.05, and a tendency to differ was declared when 0.10 ≥ P > 0.05.
Results
Differentially expressed genes
Principal component analysis of all microarray data was performed to examine the correlation and variation among the chips, revealing a total variance of 30.9% (S2 Fig). The first principal component (PC #1, x-axis) included genes with a median degree of variance (12.3%), whereas PC #2 (y-axis) and PC #3 (z-axis) encompassed genes that had low ranges of variance (9.84% and 8.75%, respectively). Overall, PCA clearly demonstrated that the chips within each treatment group were clustered closely together.
Individual ANOVA was conducted to identify altered expression of RNA transcripts in the pituitary tissue of HE vs. LE steers. At the P < 0.01 level and a false discovery rate of < 16%, 1,715 gene transcripts were identified. To refine this analysis, genes with the criteria of a false discovery rate of less than 4.8% and P < 0.001 were considered to be DEGs (S2 Table). Of these 542 DEGs, 227 (10 non-annotated) were up-regulated, 5.5% to 79.8%, and 315 (14 non-annotated) were down-regulated, 5.7% to 69.0%, in HE vs. LE steers.
Hierarchical cluster analysis of the 542 DEGs revealed that all steers were clearly separated into either the LE or HE treatment group (S3 Fig). Relative to LE steers, approximately 40% of the genes in the HE steers were up-regulated and 60% down-regulated.
Functional, canonical pathway, and gene network analyses
To determine the physiological significance of HE-induced DEGs (S2 Table), bioinformatic analysis of canonical, functional, and network pathway analyses was performed. Canonical pathway analysis revealed (P < 0.001) that the top 7 pathways were the following: axonal guidance signaling (26 genes), role of NFAT in cardiac hypertrophy (16 genes), P2Y purigenic receptor signaling pathway (13 genes), cardiac hypertrophy signaling (17 genes), Tec kinase signaling (14 genes), ErbB signaling (10 genes), and CXCR4 signaling (13 genes) (Table 1). Additionally, several affected pathways central to prolactin production, secretion, or signaling were identified (Table 2), including dopamine receptor signaling, Gαi signaling, cAMP–mediated signaling, protein kinase A signaling, and prolactin signaling. Moreover, canonical pathway analysis also identified affected pathways involved in the signaling of other pituitary-derived hormones (Table 3): corticotropin-releasing hormone signaling, melanocyte development and pigmentation signaling, growth hormone signaling, and GnRH signaling.
Table 1. Top seven IPA-identified canonical pathways of genes differentially expressed by pituitary tissue of steers grazing high (HE) vs. low (LE) endophyte-infected forages.
| Canonical Pathway | Number1 | Gene Symbol | Ratio2 | -log (P-value) |
|---|---|---|---|---|
| Axonal Guidance Signaling | 26 | ITSN1,BDNF,PIK3R1,UNC5B,GNB5,ABLIM1,SEMA4C,PLCD1,GNB4,SRGAP2,ABLIM2,ACE,GNG12,PRKCA,EPHA7,PLXNC1,PRKCQ,FES,PAK6,FGFR1,ITGA2,GNG3,PLCL2,WIPF1,SEMA3C,PRKAR1A | 0.06 | 5.38 |
| Role of NFAT in Cardiac Hypertrophy | 16 |
MAP2K6,PRKCQ,FGFR1,PIK3R1,SLC8A3,GNB5,GNG3,PLCL2,PLCD1,GNB4,MAPK10,RCAN3,ADCY8,GNG12,PRKAR1A,PRKCA |
0.08 | 5.33 |
| P2Y Purigenic Receptor Signaling Pathway |
13 |
PRKCQ,FGFR1,PIK3R1,CREB3,GNB5,GNG3,PLCL2,PLCD1,GNB4,ADCY8,GNG12,PRKCA,PRKAR1A |
0.09 | 5.04 |
| Cardiac Hypertrophy Signaling | 17 | MAP2K6,DIRAS3,PIK3R1,FGFR1,IL6R,GNB5,GNG3,MAP3K5,PLCL2,PLCD1,GNB4,RHOQ,MAPK10,ADCY8,MAP3K3,GNG12,PRKAR1A | 0.07 | 4.80 |
| Tec Kinase Signaling | 14 | GNB4,PRKCQ,RHOQ,PAK6,DIRAS3,PIK3R1,FGFR1,ITGA2,GNB5,MAPK10,GNG3,FRK,GNG12,PRKCA | 0.08 | 4.77 |
| ErbB Signaling | 10 | GNB4,PRKCQ,RHOQ,PAK6,DIRAS3,PIK3R1,FGFR1,GNB5,MAPK10,GNG3,ADCY8,GNG12,PRKCA | 0.10 | 4.41 |
| CXCR4 Signaling | 13 | GNB4,PRKCQ,RHOQ,PAK6,DIRAS3,PIK3R1,FGFR1,GNB5,MAPK10,GNG3,ADCY8,GNG12,PRKCA | 0.08 | 4.14 |
1The number of genes (listed in the “Symbol” column) associated with the particular canonical pathway.
2The ratio is calculated as the number of genes in a given pathway that meet cutoff criteria (e.g., the ANOVA P-value for the differential expression between HE and LE groups is less than 0.001) divided by the total number of genes that make up that pathway.
Table 2. IPA-identified canonical pathways of genes central to prolactin production, secretion, or signaling differentially-expressed by pituitary tissue of steers grazing high (HE) vs. low (LE) endophyte-infected forages.
| Canonical Pathway | Number1 | Gene Symbol | Ratio2 | -log (P-value) |
|---|---|---|---|---|
| Dopamine Receptor Signaling | 4 | PRL,ADCY8,DRD2,PRKAR1A | 0.04 | 0.91 |
| Gαi Signaling |
9 |
GABBR2,GNB4,GNB5,HTR1F,GNG3,ADCY8,DRD2,GNG12,PRKAR1A |
0.07 |
2.95 |
| cAMP-mediated Signaling | 9 | PDE8A,GABBR2,PKIB,CREB3,HTR1F,ADCY8,DRD2,CNGA3,PRKAR1A |
0.04 |
1.31 |
| Protein Kinase A Signaling |
20 |
PRKCQ,PTPRD,CREB3,MYLK3,GNB5,GNG3,PLCL2,CNGA3,PDE8A,PLCD1,GNB4,DUSP10,ADCY8,PTPRN,EYA1,KDELR2,GNG12,TCF7L2,PRKCA,PRKAR1A |
0.05 |
3.45 |
| Prolactin Signaling |
6 | PRKCQ,PRL,PIK3R1,FGFR1,PRLR,PRKCA | 0.07 | 2.04 |
1The number of genes (listed in the “Symbol” column) associated with the particular canonical pathway.
2The ratio is calculated as the number of genes in a given pathway that meet cutoff criteria (e.g., the ANOVA P-value for the differential expression between HE and LE groups is < 0.001) divided by the total number of genes that make up that pathway.
Table 3. IPA-identified canonical pathways of genes involved in signaling of selected pituitary-derived hormones differentially-expressed by pituitary tissue of steers grazing high (HE) vs. low (LE) endophyte-infected forages.
| Canonical Pathway | Number1 | Gene Symbol | Ratio2 | -log (P-value) |
|---|---|---|---|---|
| Melanocyte Development and Pigmentation Signaling |
7 | PIK3R1,FGFR1,CREB3,POMC,RPS6KA5,ADCY8,PRKAR1A | 0.07 | 2.38 |
| Corticotropin-releasing Hormone Signaling |
7 | PRKCQ,BDNF,CREB3,POMC,ADCY8,PRKCA,PRKAR1A | 0.06 | 1.87 |
| Growth Hormone Signaling | 6 | IGF2,PRKCQ,PIK3R1,FGFR1,RPS6KA5,PRKCA | 0.07 | 2.06 |
| GnRH Signaling | 10 | MAP2K6,PRKCQ,PAK6,CREB3,MAPK10,MAP3K5,ADCY8,MAP3K3,PRKCA,PRKAR1A | 0.07 | 3.26 |
1The number of genes (listed in the “Symbol” column) associated with the particular canonical pathway.
2The ratio is calculated as the number of genes in a given pathway that meet cutoff criteria (e.g., the ANOVA P-value for the differential expression between HE and LE groups is less than 0.001) divided by the total number of genes that make up that pathway.
To refine this analysis to pituitary-specific metabolism, IPA analysis was re-run after applying the pituitary gland-specific filter. Diseases and Bio Function Analysis found (P ≤ 0.01) putative changes in diseases and disorders, molecular and cellular functions, and physiological system development and function, resulting from the differential expression of 5 genes (DRD2, PRL, ESR1, POMC, and TCF7L2).
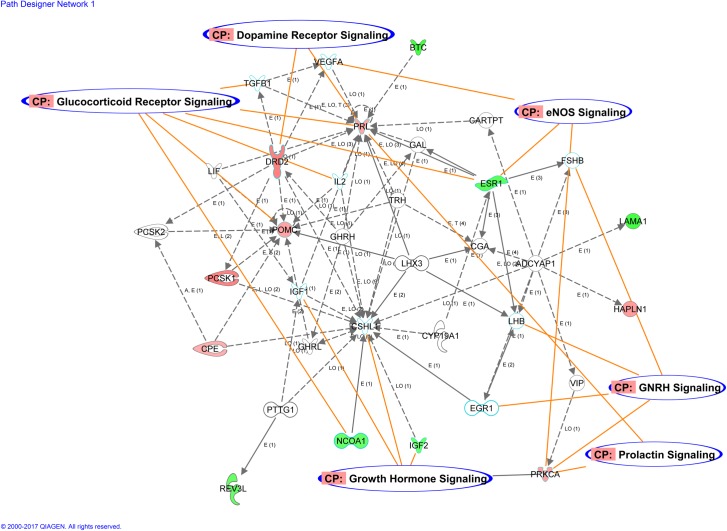
To gain insight into potentially interacting canonical pathways, pathway network analysis revealed one network that included 13 DEGs (BTC, CPE, DRD2, ESR1, HAPLN1, IGF2, LAMA1, NCOA1, PCSK1, POMC, PRKCA, PRL, and REV3L). Overlaying of canonical pathways revealed cross talk among several cell signaling pathways (Fig 1), including glucocorticoid receptor signaling (ESR1, IL2, NCOA1, POMC, PRL, TGFB1), GnRH signaling (EGR1, FSHB, LHB, PRKCA), growth hormone signaling (CSHL1, IGF1, IGF2, PRKCA), eNOS signaling (ESR1, PRKCA, VEGFA), dopamine receptor signaling (DRD2, PRL), and prolactin signaling (PRKCA, PRL).
Fig 1. Canonical pathway network analysis.
The red or green coloring represents down- or up-regulation, respectively, whereas no color indicates the molecule was added from the Ingenuity Knowledge Base (Ingenuity Pathway, Ingenuity Systems, Inc., Redwood City, CA). The intensity of the node color (light to dark) proportionally indicates the degree of differential expression. Straight lines represent binding only, whereas arrowheads symbolize action-on. A crosshead bar signifies inhibition. Labels of interaction or relationship: A = Activation, CP = Canonical Pathway, E = Expression (includes metabolism or synthesis for chemicals), I = Inhibition, LO = Localization. The number in parenthesis for each interaction indicates the number of published references in the Ingenuity Knowledge Base that support the particular interaction.
Real-time reversed-transcribed PCR analysis of selected mRNA
Real-time RT-PCR analysis was used to corroborate the microarray analysis-identified altered expression of key genes responsible for prolactin synthesis and secretion and POMC/ACTH production in HE vs. LE steers (Table 4). The results of these two analyses were consistent for all the targeted genes, with the exception of PRLR, although the statistical significance (ANOVA P-value) and fold changes measured by the two analytical techniques differed for some genes. For PRLR, unlike the microarray analysis, RT-PCR analysis was designed to delineate the long form (l-PRLR) and short form (s-PRLR). In the microarray analysis, PRLR was down-regulated in HE steers (P < 0.001), whereas in RT-PCR analysis, expression of s-PRLR was not altered (P = 0.21) and expression of l-PRLR had a tendency to differ (P < 0.07) in HE vs. LE steers.
Table 4. Comparison of microarray and real-time RT-PCR identification of selected genes by pituitary tissue of steers grazing high (HE) vs. low (LE) endophyte-infected forages.
| Gene | Gene Name | Microarray | Real-time RT-PCR | ||||
|---|---|---|---|---|---|---|---|
| Change2 | Ratio3 | P-value | Change2 | Ratio3 | P-value | ||
| ACTB1 | Actin, beta | 1.03 | 1.03 | 0.084 | 1.01 | 1.01 | 0.568 |
| PPIA1 | Peptidylprolyl isomerase A | -1.07 | 0.93 | 0.441 | 1.00 | 1.00 | 0.422 |
| UBC1 | Ubiquitin C | 1.00 | 1.00 | 0.994 | 1.00 | 1.00 | 0.816 |
| DRD2 | Dopamine receptor D2 | -1.76 | 0.57 | 0.001 | -2.14 | 0.47 | 0.001 |
| PRL | Prolactin | -1.23 | 0.81 | 0.001 | -5.67 | 0.18 | 0.001 |
| PRLR | Prolactin receptor | -1.31 | 0.76 | 0.001 | NA | NA | NA |
| s-PRLR | Prolactin receptor short isoform | NA | NA | NA | -1.20 | 0.83 | 0.210 |
| l- PRLR | Prolactin receptor long isoform | NA | NA | NA | -1.29 | 0.78 | 0.062 |
| POU1F1 | POU class 1 homeobox 1 | -1.30 | 0.77 | 0.003 | -1.47 | 0.68 | 0.038 |
| GAL | Galanin/GMAP prepropeptide | -1.34 | 0.74 | 0.009 | -2.35 | 0.43 | 0.019 |
| VIP | Vasoactive intestinal peptide | -1.76 | 0.57 | 0.003 | -2.08 | 0.48 | 0.045 |
| POMC | Proopiomelanocortin | -1.25 | 0.80 | 0.001 | -2.27 | 0.44 | 0.006 |
| PCSK1 | Proprotein convertase subtilisin/kexin type 1 | -1.72 | 0.58 | 0.001 | -2.02 | 0.5 | 0.001 |
| GH1 | Growth Hormone 1 | 1.01 | 1.01 | 0.728 | 1.20 | 1.20 | 0.436 |
| TSHB | Thyroid stimulating hormone beta | 1.00 | 1.00 | 0.999 | 1.14 | 1.14 | 0.418 |
| TBX19 | T-Box 19 | -1.14 | 0.88 | 0.104 | -1.20 | 0.84 | 0.217 |
| NeuroD1 | Neuronal differentiation 1 | 1.19 | 1.19 | 0.178 | 1.18 | 1.18 | 0.415 |
| NR3C1 | Nuclear receptor subfamily 3 group C member 1 | 1.14 | 1.14 | 0.112 | 1.33 | 1.33 | 0.270 |
| CRHR1 | Corticotropin-releasing hormone receptor 1 | 1.19 | 1.19 | 0.106 | 1.39 | 1.39 | 0.192 |
1Expression reference genes.
2Data are expressed as fold change in HE relative to LE expression.
3Data are expressed as ratio of HE relative to LE expression.
Although microarray analysis did not identify them as DEGs (S3 Table), the expression of 3 genes was assessed by RT-PCR analyses because they are known targets of POU1F1 (GH1, TSHB) or are involved in CRH stimulation of ACTH production (CRHR1). RT-PCR analysis corroborated the microarray analysis that pituitary expression of these genes did not differ between HE and LE steers (Table 4).
Differentially expressed miRNAs (DEMs) and their predicted target genes associated with prolactin and POMC/ACTH production
The microarray chips used for this study detected 574 miRNAs. Of these, only 6 were differentially expressed (P < 0.001) in HE vs. LE steers (S2 Table). Specifically, miR-380 (42%), miR-2318 (17%), miR-329B (36%), and miR-544A (38%) were down-regulated in HE vs. LE steers, whereas miR-2356 (38%) and miR-2400 (8%) were up-regulated. The target genes of these DEMs that were associated with prolactin or POMC/ACTH production are listed in Table 5. Although no miRNAs known to directly target mRNA for prolactin were differentially expressed, every DEM targeted multiple prolactin transcription factors, stimulators, and (or) inhibitors, including miR-544A that targeted all the PRL-associated genes. Overall, the mRNA for three transcription factors (POULF1, ESR1, PREB), two transcription stimulators (EGF, IKZF1), and one transcription inhibitor (PKIA) of PRL were predicted to be targets of the DEMs. With specific regard to microarray-identified DEGs (Table 4) targeted by DEMs, PRLR was predicted to be the target of five DEMs and ESR1 the target of four DEMs.
Table 5. Predicted relationship between differentially-expressed mRNA of prolactin and ACTH pathway genes, including transcription factors (TF), transcription stimulators (TS), and transcription inhibitors (TI), known to be targets of microarray-identified differentially-expressed miRNAs (DEMs)1.
| Gene Symbol | Gene Description | DEM (P < 0.001)2,3 |
|---|---|---|
| PRL | Prolactin | |
| PRLR | Prolactin Receptor | miR-329B, miR-380, miR-544A, miR-2318, miR-2356 |
| DRD2 | Dopamine Receptor D2 | |
| POU1F1 (TF for PRL) | POU Class 1 Homeobox 1 | miR-544A |
| VIP | Vasoactive Intestinal Peptide | miR-544A, miR-2400 |
| ESR1 (TF for PRL) | Estrogen Receptor 1 | miR-329B, miR-380, miR-544A, miR-2356 |
| PREB (TF for PRL) | Prolactin Regulatory Element Binding | miR-544A, miR-2400 |
| EGF (TS for PRL) | Epidermal Growth Factor | miR-380, miR-544A, miR-2356 |
| IKZF1 (TS for PRL) | IKAROS Family Zinc Finger 1 | miR-380, miR-544A, miR-2400 |
| PKIA (TI for PRL) | CAMP-Dependent Protein Kinase Inhibitor Alpha | miR-329B, miR-380, miR-544A, miR-2318, miR-2356, miR-2400 |
| POMC | Proopiomelanocortin | |
| PCSK1 | Proprotein Convertase Subtilisin/Kexin Type 1 | miR-380 |
| TBX19 (TF for POMC) | T-Box 19 | miR-380 |
| NEUROD1 (TF for POMC) | Neuronal Differentiation 1 | miR-380, miR-544A, miR-2318 |
| JUN (TF for POMC) | Jun Proto-Oncogene, AP-1 Transcription Factor Subunit | miR-2400 |
| LEP (TS for POMC) | Leptin | miR-544A |
| LIF (TS for POMC) | Leukemia Inhibitory Factor | miR-2400 |
| NR3C1 (TI for POMC) | Nuclear Receptor Subfamily 3 Group C Member 1 | miR-380, miR-544A, miR-2318, miR-2356 |
| SMARCA4 (TI for POMC) | SWI/SNF Related, Matrix Associated, Actin Dependent Regulator Of Chromatin, Subfamily A, Member 4 | miR-329B |
| HDAC2 (TI for POMC) | Histone Deacetylase 2 | miR-380, miR-2356 |
1Putative gene targets of DEMs were identified using TargetScan (Release 7.1, http://www.targetscan.org).
2miR-329B: GenBank accession number is NR_031209 and is known as miR-329 by TargetScan.
3miR-544A: GenBank accession number is NR_031187 and is known as miR-544 by TargetScan.
Analogously, for POMC/ACTH production genes, whereas no miRNAs were differentially expressed that targeted POMC per se, TargetScan predicted that DEMs would interact with the mRNA of three transcription factors (TBX19, NEUROD1, JUN), two transcription stimulators (LEP, LIF), and three transcription inhibitors (NR3C1, SMARCA4, HDAC2) of the POMC production pathway. With specific regard to microarray-identified DEGs targeted by DEMs, PCSK1 was the target of a single miRNA (miR-380).
Because POMC expression was altered and miR-380 is predicted to target two POMC transcription factors (NEUROD1, TBX19), the expression of NEUROD 1 and TBX19 was evaluated by RT-PCR, although their expression was not altered as determined by microarray analysis (S3 Table). However, consistent with the microarray analysis, the expression of NEUROD 1 and TBX19 based on RT-PCR analysis was not altered (Table 4).
Although the expression of NR3C1 was not affected based on microarray analysis (S3 Table), the expression was evaluated by RT-PCR because glucocorticoid receptor complex represses the POMC gene through a negative glucocorticoid response element of POMC promoter [17]. However, RT-PCR analysis found no difference in NR3C1 abundance in the pituitaries of HE and LE steers (Table 4).
Discussion
The pituitary is an endocrine gland composed of anterior, intermediate, and posterior lobes, with the anterior lobe occupying approximately 80% of the entire gland. The anterior lobe is composed of five tropic cell types, which together secrete six hormones: corticotrophs (ACTH), gonadotrophs (FSH and LH), lactotrophs (prolactin), somatotrophs (GH), and thyrotrophs (TSH). Previous studies show that hormone production by all five anterior pituitary cell types is affected by the consumption of ergot alkaloids in cattle [18, 19], with decreased concentrations of serum prolactin one of the most common serological signs [20, 21].
To our knowledge, the effect of ergot alkaloid consumption on pituitary transcriptomic profiles has not been reported. To obtain this information, we conducted transcriptome analysis of pituitaries collected from previously described [5] beef steers suffering from fescue toxicosis induced by summer-long grazing (89 to 105 d) of HE and LE pastures. Importantly, concentrations of prolactin in the serum of HE steers were only approximately 10% of those of the LE steers [5], and the glucocorticoid receptor-mediated pathway was implicated in observed changes in carbohydrate metabolism in HE steers [6]. As noted in the Introduction, the goal of the current research was to determine whether gene expression profiles differed between whole pituitaries of HE and LE steers using transcriptome and targeted gene expression analysis and to identify specific candidate molecules and signaling pathways responsible for the altered physiology of steers consuming ergot alkaloid-containing tall fescue. The global hypothesis tested was that consumption of endophyte-infected tall fescue would alter pituitary transcriptome profiles. At the P < 0.001 level, the microarray analysis approach revealed the differential expression of 542 RNA transcripts by the pituitary. Importantly, the pattern of altered gene expression was clearly dependent on treatment according to hierarchical cluster analysis (S3 Fig). Thus, the first salient finding of this study is that summer-long grazing of endophyte-infected tall fescue alters the pituitary transcriptome; thus, the global hypothesis is accepted.
More specifically, given that the serum prolactin concentrations of HE steers were only approximately 10% of those of the LE steers [5], and that the glucocorticoid receptor-mediated pathway was implicated in observed changes in carbohydrate metabolism in HE steers [6], the expectation was that the expression pattern for pituitary genes responsible for the production and secretion of prolactin would be consistent with a down-regulated capacity, whereas that for POMC/ACTH would be consistent with an up-regulated capacity. Conclusions reached about these hypotheses, as well as the possible roles of miRNA in these processes, are discussed below.
Fescue toxicosis and prolactin synthesis and secretion
The effect of ergot alkaloid consumption on prolactin production and secretion is best understood through the interactive pathway of dopamine receptors located on the surface of lactotrophs. Dopamine is one of the most influential regulators of prolactin secretion. Activation of the dopamine receptor suppresses PRL gene expression via the inhibition of adenylyl cyclase and prolactin exocytosis through modification of several potassium and calcium channels [22]. One way by which ergot alkaloid consumption directly affects lactotrophs is through the binding and stimulation of dopamine type two receptors (DRD2) on the cell surface [22]. Ergot alkaloids ingested with consumption of endophyte-infested tall fescue structurally resemble various biogenic amines, such as dopamine [3]. These ergot amines can bind to dopamine type two receptors, stimulate the receptors, and reduce basal level prolactin production and secretion as described above [23]. Consistent with this understanding, the HE steers in this study had serum prolactin concentrations that were only 10% of those of the LE steers [5]. The lower prolactin found in serum of steers exposed to HE pasture directly corresponded to the microarray and real-time RT-PCR results regarding the gene expression of DRD2, POU1F1 (a.k.a. Pit1), PRL and PRLR genes (Table 4). Based on real-time RT-PCR results, the expression of these genes decreased by approximately 53%, 32%, 82%, and 22% (long isoform of prolactin receptor with tendency to differ), respectively, in HE vs. LE steers. POU1F1 plays a pivotal role in PRL expression by binding to specific sites of promoter elements in the PRL gene [24]. Therefore, decreased expression of POU1F1 might explain reduced PRL mRNA expression in HE steers to a certain extent.
An apparently associated finding was the accompanying down regulation of both DRD2 and PRLR genes. Although speculative, a decrease in the expression of DRD2 may have been a preventive measure by the lactotrophic cells to counteract the suppression of prolactin production due to the activation of the dopamine receptors, whereas the down regulation of prolactin receptor mRNA in pituitary tissue may be the result of a decreased requirement for prolactin binding in a prolactin-poor environment. Additionally, expression of GAL (galanin/GMAP prepropeptide) and VIP (vasoactive intestinal peptide) also decreased in HE vs. LE steers according to both microarray and real-time RT-PCR results (Table 4). Galanin is known to stimulate prolactin release [25, 26], although the mechanism has not been clearly defined. Additionally, galanin may directly stimulate prolactin expression and act as a lactotroph growth factor, particularly when exposure to estrogen is high [26]. Vasoactive intestinal peptide also stimulates prolactin secretion in multiple species, with receptors found on lactotrophs [27–30]. Although the mechanism by which vasoactive intestinal peptide stimulates prolactin release is not well delineated, as for galanin, cAMP accumulation and a delayed increase in calcium concentration were observed in the process [30, 31]. Thus, our hypothesis that at least the pituitary genes responsible for the production and secretion of prolactin would be down-regulated is also accepted.
In addition to prolactin, POU1F1 activates growth hormone (GH1) promoter transcriptionally [32] and is involved in thyrotropin-releasing hormone (TRH) stimulation of the beta subunit of thyroid-stimulating hormone (TSHB) expression [33]. However, RT-PCR (Table 4) analysis corroborated the microarray (S3 Table) findings that neither GH1 nor TSHB was differentially expressed in HE vs. LE steers.
Although best known for the role in regulating lactation, prolactin affects a wide variety of biological functions [34, 35], including reproduction, osmoregulation, antiangiogenic activity, regulation of immune responses, regulation of insulin release, and control of growth. With regard to growth, prolactin is associated with food intake and body weight and may interact with hypothalamic neurons responsible for appetite regulation [36, 37]. Moreover, as described in detail previously [5], the average daily gain of HE steers was 31% less than that of LE steers (P < 0.05), and the final body weight of HE steers was 7.4% less than that of LE steers (P < 0.05). Hence, reduced prolactin concentrations in HE steers might account for these observations to a certain degree.
Fescue toxicosis, POMC/ACTH synthesis, and gluconeogenesis
As noted in the Introduction, increased mitochondrial mass and capacity for ATP synthesis and amino acid-derived gluconeogenesis [5] are postulated to be coordinated through the glucocorticoid receptor-mediated pathway [6]. Therefore, a reasonable hypothesis is that the capacity for glucocorticoid synthesis (POMC/ACTH production) would be elevated in the pituitaries of HE vs. LE steers. However, although we did not measure ACTH stimulation of cortisol release by the adrenal glands, canonical pathway analysis of pituitary DEGs indicated (z-score less than -2.00) the down-regulation of the corticotropin-releasing hormone (CRH) signaling pathway (BDNF, POMC, ADCY8, PRKCA, and PRKAR1A) in HE steers (Table 3). As part of the hypothalamic–pituitary–adrenal axis, the primary function of CRH is to stimulate ACTH production from the pituitary through interaction with CRHR1, the predominant pituitary-expressed CRH receptor. According to the microarray and RT-PCR analyses (S3 Table, Table 4), CRHR1 mRNA expression level was not affected in HE vs. LE steers, whereas CRHR2 was not qualified for RT-PCR analysis because of the low expression level. These findings are consistent with the understanding that CRHR1 is highly expressed by the pituitary, whereas CRHR2 is predominately expressed by brain and peripheral tissues [38]. ACTH is synthesized within the anterior pituitary as part of the much larger precursor molecule proopiomelanocortin (POMC), which is cleaved into smaller peptide hormones in a tissue-specific manner by proprotein convertases. In pituitary corticotrophs, proprotein convertase 1 (encoded by the PCSK1 gene) alone is expressed and cleaves POMC, producing ACTH, β-endorphin, β-lipotrophin, amino-terminal peptide, and joining peptide [39]. According to the microarray and real-time RT-PCR analyses (Table 4), the abundance of both POMC and PCSK1 mRNA was reduced in the pituitaries of HE vs. LE steers. Thus, the hypothesis that expression of pituitary genes responsible for the production of POMC/ACTH would be increased is rejected.
Despite the importance to adrenal steroidogenesis, research describing the effects of fescue toxicosis on blood ACTH is lacking. Moreover, although studies have been conducted to better understand the relationship between fescue toxicosis and circulating cortisol in cattle, their results are discordant [40–42]. To resolve the apparent enigma that HE steers displayed a reduced potential for pituitary synthesis of ACTH (this study), yet increased hepatic gluconeogenesis capacity [5, 6], further research is required.
Role of miRNAs in regulating prolactin and POMC/ACTH pathways
Messenger RNA abundance is regulated by a combination of pre-transcription and post-transcription events. Transcription factors contribute to mRNA abundance at the pre-transcription level by binding to DNA and either positively or negatively regulating gene transcription [43]. MicroRNAs regulate mRNA abundance at the post-transcriptional level through complementary binding of target mRNA transcripts, resulting in repressed translation or enhanced degradation of bound mRNA [44]. Thus, decreased expression of a given miRNA would result in increased target mRNA abundance and vice versa. miR-544A, which putatively regulates multiple transcription factors and stimulators (ESR1 [45], EGF [46], IKZF1 [47], POU1F1 [48], PREB [49], VIP [50]) of the prolactin gene (Table 5), was down-regulated 38% in HE vs. LE steers; however, expression of PRL decreased in HE vs. LE steers. Inconsistency between the abundance of miR-380 and its target gene was also found. Because miR-380 is predicted to target PCSK1 and two POMC transcription factors, NEUROD1 [51] and TBX19 [52] (Table 5), we expected the decrease in expression of miR-380 (42%) in the pituitaries of HE vs. LE steers to result in increased expression of POMC and PCSK1. However, microarray (S3 Table) and RT-PCR (Table 4) results showed no difference in expression level of NEUROD1 and TBX19 mRNA in pituitaries of HE vs. LE steers, whereas both POMC and PCSK1were down-regulated.
Although evidence shows that miRNAs can also up-regulate gene expression [53], an alternative explanation to the above inconsistencies could be due to the stringency level of P-values that was applied to the microarray analysis. Any given gene is usually regulated by numerous miRNAs, and the complements of these miRNAs decide the fate of the transcription of the gene. Thus, a stringent significant cutoff criterion (P < 0.001) could filter out potential miRNAs targeting genes of interest. For example, one striking finding listed in Table 5 is that no DEMs were identified that targeted DRD2, PRL, or POMC. However, when the P-value was relaxed to 0.05, then multiple miRNAs predicted to target DRD2 (miR-141, miR-214, miR-584, miR-631, miR-2316, miR-2350, miR-2373, miR-2382, miR-2418, miR-2464) were identified. Additionally, miR-2335 and miR-2399 (predicted to target both VIP and its transcription factor NURR1 [54] (encoded by NR4A2)) also became candidate regulators of PRL expression. Collectively, the evidence suggested that altered expression of miRNAs might have affected mRNA abundance by affecting both pre- and post-transcription events of genes regulating prolactin and POMC/ACTH pathways.
This experiment is part of a comprehensive study to understand the whole body and tissue-specific effects of ergot alkaloid consumption in cattle [5, 6, 55]. The unique pituitary-specific findings of this study are an important contribution to our understanding of how ergot alkaloids exert their deleterious effects on cattle production. In summary, the findings indicate that anterior pituitary functions were globally impaired in steers consuming high-toxic endophyte-infected tall fescue. In addition to inhibiting the abilities to synthesize and secrete prolactin (a function of lactotrophs), ACTH synthesis capacity (a function of corticotrophs) might have been reduced. Canonical pathway analysis also indicated that growth hormone signaling and GnRH signaling were altered in HE vs. LE steers (Table 3). A larger implication of this research may be that it allows for selective breeding for genotypes with a higher resistance to endophyte toxicosis, because the specific genes and networks of genes have now been identified that are susceptible to ergot alkaloids contained in endophyte-infected tall fescue. Likewise, with the identification of putative ergot alkaloid sensitive mechanisms within the pituitary gland, this new knowledge may help to develop dietary treatments that ameliorate the effects of ergot alkaloid ingestion [7].
Supporting information
Within a sequence, underlined nucleotides indicate the forward and reverse primer positions.
(DOCX)
The red and blue dots represent linear combinations of the relative expression data, including expression values and variances, of the 26,675 gene transcripts in each Bovine GeneChip.
(DOCX)
As indicated by the legend color box, white color in the middle represents the mean value, 0; red color represents gene expression levels above the mean expression; and blue color denotes expression below the mean. The intensity of the color reflects the relative intensity of the fold change.
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
This is publication No. 17-07-076 of the Kentucky Agricultural Experiment Station and is published with approval of the Director. This work is supported by a United States Department of Agriculture-Agricultural Research Service Cooperative Agreement (JCM) and by the National Institute of Food and Agriculture, U.S. Department of Agriculture, Hatch project No. KY007095 (JCM, PJB).
Data Availability
All relevant data are within the paper and its Supporting Information files, except for the .cel files which are available from GEO (http://www.ncbi.nlm.nih.gov/geo/) as accession no. GSE62570.
Funding Statement
This work is supported by a United States Department of Agriculture-Agricultural Research Service Cooperative Agreement (JCM) and by the National Institute of Food and Agriculture, U.S. Department of Agriculture, Hatch project No. KY007095 (JCM, PB). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Aiken G, Strickland J. Forages and pastures symposium: managing the tall fescue–fungal endophyte symbiosis for optimum forage-animal production. Journal of animal science. 2013;91(5):2369–78. doi: 10.2527/jas.2012-5948 [DOI] [PubMed] [Google Scholar]
- 2.Siegel MR, Bush LP. Importance of endophytes in forage grasses, a statement of problems and selection of endophytes. Biotechnology of endophytic fungi of grasses. 1994. [Google Scholar]
- 3.Strickland JR, Looper ML, Matthews J, Rosenkrans C, Flythe M, Brown K. Board-invited review: St. Anthony’s Fire in livestock: causes, mechanisms, and potential solutions. Journal of animal science. 2011;89(5):1603–26. doi: 10.2527/jas.2010-3478 [DOI] [PubMed] [Google Scholar]
- 4.Beardwell C, Robertson GL. The Pituitary. Beardwell C, Robertson GL, editors. London; Boston: Butterworths; 1981. 337 p. [Google Scholar]
- 5.Brown K, Anderson G, Son K, Rentfrow G, Bush L, Klotz J, et al. Growing steers grazing high versus low endophyte (Neotyphodium coenophialum)-infected tall fescue have reduced serum enzymes, increased hepatic glucogenic enzymes, and reduced liver and carcass mass. Journal of animal science. 2009;87(2):748–60. doi: 10.2527/jas.2008-1108 [DOI] [PubMed] [Google Scholar]
- 6.Liao SF, Boling JA, Matthews JC. Gene expression profiling indicates an increased capacity for proline, serine, and ATP synthesis and mitochondrial mass by the liver of steers grazing high vs. low endophyte-infected tall fescue. Journal of animal science. 2015;93(12):5659–71. doi: 10.2527/jas.2015-9193 . [DOI] [PubMed] [Google Scholar]
- 7.Matthews J, Bridges P. NutriPhysioGenomics applications to identify adaptations of cattle to consumption of ergot alkaloids and inorganic versus organic forms of selenium: altered nutritional, physiological and health states? Animal Production Science. 2014;54(10):1594–604. [Google Scholar]
- 8.Matthews JC, Zhang Z, Patterson JD, Bridges PJ, Stromberg AJ, Boling J. Hepatic transcriptome profiles differ among maturing beef heifers supplemented with inorganic, organic, or mixed (50% inorganic: 50% organic) forms of dietary selenium. Biological trace element research. 2014;160(3):321–39. doi: 10.1007/s12011-014-0050-4 [DOI] [PubMed] [Google Scholar]
- 9.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–64. doi: 10.1093/biostatistics/4.2.249 . [DOI] [PubMed] [Google Scholar]
- 10.Wu Z, Irizarry RA, Gentleman R, Martinez-Murillo F, Spencer F. A model-based background adjustment for oligonucleotide expression arrays. Journal of the American statistical Association. 2004;99(468):909–17. [Google Scholar]
- 11.Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, et al. Minimum information about a microarray experiment (MIAME)—toward standards for microarray data. Nature genetics. 2001;29(4):365–71. doi: 10.1038/ng1201-365 [DOI] [PubMed] [Google Scholar]
- 12.Bridges PJ, Jeoung M, Shim S, Park JY, Lee JE, Sapsford LA, et al. Hematopoetic prostaglandin D synthase: an ESR1-dependent oviductal epithelial cell synthase. Endocrinology. 2012;153(4):1925–35. doi: 10.1210/en.2011-1900 ; PubMed Central PMCID: PMCPMC3320253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. methods. 2001;25(4):402–8. doi: 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 14.Lisowski P, Pierzchala M, Goscik J, Pareek CS, Zwierzchowski L. Evaluation of reference genes for studies of gene expression in the bovine liver, kidney, pituitary, and thyroid. J Appl Genet. 2008;49(4):367–72. doi: 10.1007/BF03195635 . [DOI] [PubMed] [Google Scholar]
- 15.Thompson I, Ozawa M, Bubolz J, Yang Q, Dahl G. Bovine luteal prolactin receptor expression: potential involvement in regulation of progesterone during the estrous cycle and pregnancy. Journal of animal science. 2011;89(5):1338–46. doi: 10.2527/jas.2010-3559 [DOI] [PubMed] [Google Scholar]
- 16.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4:e05005 doi: 10.7554/eLife.05005 ; PubMed Central PMCID: PMCPMC4532895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Drouin J, Sun Y, Chamberland M, Gauthier Y, De Lean A, Nemer M, et al. Novel glucocorticoid receptor complex with DNA element of the hormone-repressed POMC gene. The EMBO Journal. 1993;12(1):145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Browning R Jr., Leite-Browning ML, Smith HM, Wakefield T Jr. Effect of ergotamine and ergonovine on plasma concentrations of thyroid hormones and cortisol in cattle. Journal of animal science. 1998;76(6):1644–50. . [DOI] [PubMed] [Google Scholar]
- 19.Browning R, Thompson F, Sartin J, Leite-Browning M. Plasma concentrations of prolactin, growth hormone, and luteinizing hormone in steers administered ergotamine or ergonovine. Journal of animal science. 1997;75(3):796–802. [DOI] [PubMed] [Google Scholar]
- 20.Hurley W, Convey E, Leung K, Edgerton L, Hemken R. Bovine Prolactin, TSH, T and T Concentrations as Affected by Tall Fescue Summer Toxicosis and Temperature. Journal of animal science. 1980;51(2):374–9. [DOI] [PubMed] [Google Scholar]
- 21.Lipham LB, Thompson FN, Stuedemann JA, Sartin JL. Effects of metoclopramide on steers grazing endophyte-infected fescue. Journal of animal science. 1989;67(4):1090–7. . [DOI] [PubMed] [Google Scholar]
- 22.Fitzgerald P, Dinan TG. Prolactin and dopamine: what is the connection? A review article. Journal of psychopharmacology (Oxford, England). 2008;22(2 Suppl):12–9. Epub 2008/08/16. doi: 10.1177/0269216307087148 . [DOI] [PubMed] [Google Scholar]
- 23.Larson BT, Harmon DL, Piper EL, Griffis LM, Bush LP. Alkaloid binding and activation of D2 dopamine receptors in cell culture. Journal of animal science. 1999;77(4):942–7. Epub 1999/05/18. . [DOI] [PubMed] [Google Scholar]
- 24.Fox SR, Jong MT, Casanova J, Ye Z-S, Stanley F, Samuels HH. The homeodomain protein, Pit-1/GHF-1, is capable of binding to and activating cell-specific elements of both the growth hormone and prolactin gene promoters. Molecular Endocrinology. 1990;4(7):1069–80. doi: 10.1210/mend-4-7-1069 [DOI] [PubMed] [Google Scholar]
- 25.Koshiyama H, Kato Y, Inoue T, Murakami Y, Ishikawa Y, Yanaihara N, et al. Central galanin stimulates pituitary prolactin secretion in rats: possible involvement of hypothalamic vasoactive intestinal polypeptide. Neurosci Lett. 1987;75(1):49–54. . [DOI] [PubMed] [Google Scholar]
- 26.Wynick D, Small CJ, Bacon A, Holmes FE, Norman M, Ormandy CJ, et al. Galanin regulates prolactin release and lactotroph proliferation. Proceedings of the National Academy of Sciences. 1998;95(21):12671–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gourdji D, Bataille D, Vauclin N, Grouselle D, Rosselin G, Tixier-Vidal A. Vasoactive intestinal peptide (VIP) stimulates prolactin (PRL) release and cAMP production in a rat pituitary cell line (GH3/B6). Additive effects of VIP and TRH on PRL release. FEBS Lett. 1979;104(1):165–8. . [DOI] [PubMed] [Google Scholar]
- 28.Frawley LS, Neill JD. Stimulation of prolactin secretion in rhesus monkeys by vasoactive intestinal polypeptide. Neuroendocrinology. 1981;33(2):79–83. . [DOI] [PubMed] [Google Scholar]
- 29.Macnamee M, Sharp P, Lea R, Sterling R, Harvey S. Evidence that vasoactive intestinal polypeptide is a physiological prolactin-releasing factor in the bantam hen. General and comparative endocrinology. 1986;62(3):470–8. [DOI] [PubMed] [Google Scholar]
- 30.Samson WK, Said SI, Snyder G, McCann SM. In vitro stimulation of prolactin release by vasoactive intestinal peptide. Peptides. 1981;1(4):325–32. [DOI] [PubMed] [Google Scholar]
- 31.Bjøro T, Østberg B, Sand O, Gordeladze J, Iversen J-G, Torjesen P, et al. Vasoactive intestinal peptide and peptide with N-terminal histidine and C-terminal isoleucine increase prolactin secretion in cultured rat pituitary cells (GE 4 C 1) via a cAMP-dependent mechanism which involves transient elevation of intracellular Ca 2+. Molecular and cellular endocrinology. 1987;49(2):119–28. [DOI] [PubMed] [Google Scholar]
- 32.Mangalam HJ, Albert VR, Ingraham HA, Kapiloff M, Wilson L, Nelson C, et al. A pituitary POU domain protein, Pit-1, activates both growth hormone and prolactin promoters transcriptionally. Genes & development. 1989;3(7):946–58. [DOI] [PubMed] [Google Scholar]
- 33.Steinfelder HJ, Hauser P, Nakayama Y, Radovick S, McClaskey JH, Taylor T, et al. Thyrotropin-releasing hormone regulation of human TSHB expression: role of a pituitary-specific transcription factor (Pit-1/GHF-1) and potential interaction with a thyroid hormone-inhibitory element. Proceedings of the National Academy of Sciences. 1991;88(8):3130–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Freeman ME, Kanyicska B, Lerant A, Nagy G. Prolactin: structure, function, and regulation of secretion. Physiological reviews. 2000;80(4):1523–631. [DOI] [PubMed] [Google Scholar]
- 35.Lamberts S, Macleod R. Regulation of prolactin secretion at the level of the lactotroph. Physiological Reviews. 1990;70(2):279–318. [DOI] [PubMed] [Google Scholar]
- 36.Ben-Jonathan N, Hugo ER, Brandebourg TD, LaPensee CR. Focus on prolactin as a metabolic hormone. Trends in Endocrinology & Metabolism. 2006;17(3):110–6. [DOI] [PubMed] [Google Scholar]
- 37.Naef L, Woodside B. Prolactin/leptin interactions in the control of food intake in rats. Endocrinology. 2007;148(12):5977–83. doi: 10.1210/en.2007-0442 [DOI] [PubMed] [Google Scholar]
- 38.Holsboer F. The rationale for corticotropin-releasing hormone receptor (CRH-R) antagonists to treat depression and anxiety. Journal of psychiatric research. 1999;33(3):181–214. [DOI] [PubMed] [Google Scholar]
- 39.Millington GW. The role of proopiomelanocortin (POMC) neurones in feeding behaviour. Nutrition & metabolism. 2007;4(1):18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aldrich CG, Paterson JA, Tate JL, Kerley MS. The effects of endophyte-infected tall fescue consumption on diet utilization and thermal regulation in cattle. Journal of animal science. 1993;71(1):164–70. . [DOI] [PubMed] [Google Scholar]
- 41.Schuenemann GM, Hockett ME, Edwards JL, Rohrbach NR, Breuel KF, Schrick FN. Embryo development and survival in beef cattle administered ergotamine tartrate to simulate fescue toxicosis. Reprod Biol. 2005;5(2):137–50. [PubMed] [Google Scholar]
- 42.Looper ML, Reiter ST, Williamson BC, Sales MA, Hallford DM, Rosenkrans CF Jr. Effects of body condition on measures of intramuscular and rump fat, endocrine factors, and calving rate of beef cows grazing common bermudagrass or endophyte-infected tall fescue. Journal of animal science. 2010;88(12):4133–41. doi: 10.2527/jas.2010-3192 . [DOI] [PubMed] [Google Scholar]
- 43.Latchman DS. Transcription factors: an overview. Int J Biochem Cell Biol. 1997;29(12):1305–12. . [DOI] [PubMed] [Google Scholar]
- 44.Engels BM, Hutvagner G. Principles and effects of microRNA-mediated post-transcriptional gene regulation. Oncogene. 2006;25(46):6163–9. doi: 10.1038/sj.onc.1209909 . [DOI] [PubMed] [Google Scholar]
- 45.Waterman ML, Adlerf S, Nelson C, Greene GL, Evans RM, Rosenfeld MG. A single domain of the estrogen receptor confers deoxyribonucleic acid binding and transcriptional activation of the rat prolactin gene. Molecular Endocrinology. 1988;2(1):14–21. doi: 10.1210/mend-2-1-14 [DOI] [PubMed] [Google Scholar]
- 46.Murdoch GH, Potter E, Nicolaisen AK, Evans RM, Rosenfeld MG. Epidermal growth factor rapidly stimulates prolactin gene transcription. Nature. 1982;300(5888):192–4. [DOI] [PubMed] [Google Scholar]
- 47.Ezzat S, Yu S, Asa SL. The zinc finger Ikaros transcription factor regulates pituitary growth hormone and prolactin gene expression through distinct effects on chromatin accessibility. Molecular Endocrinology. 2005;19(4):1004–11. doi: 10.1210/me.2004-0432 [DOI] [PubMed] [Google Scholar]
- 48.Nelson C, Albert VR, Elsholtz HP, Lu LI-W, Rosenfeld MG. Activation of cell-specific expression of rat growth hormone and prolactin genes by a common transcription factor. Science. 1988;239(4846):1400 [DOI] [PubMed] [Google Scholar]
- 49.Fliss MS, Hinkle PM, Bancroft C. Expression cloning and characterization of PREB (prolactin regulatory element binding), a novel WD motif DNA-binding protein with a capacity to regulate prolactin promoter activity. Mol Endocrinol. 1999;13(4):644–57. doi: 10.1210/mend.13.4.0260 . [DOI] [PubMed] [Google Scholar]
- 50.Bredow S, Kacsoh B, Obal F Jr., Fang J, Krueger JM. Increase of prolactin mRNA in the rat hypothalamus after intracerebroventricular injection of VIP or PACAP. Brain Res. 1994;660(2):301–8. . [DOI] [PubMed] [Google Scholar]
- 51.Poulin G, Turgeon B, Drouin J. NeuroD1/beta2 contributes to cell-specific transcription of the proopiomelanocortin gene. Molecular and cellular biology. 1997;17(11):6673–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lamonerie T, Tremblay JJ, Lanctot C, Therrien M, Gauthier Y, Drouin J. Ptx1, a bicoid-related homeo box transcription factor involved in transcription of the pro-opiomelanocortin gene. Genes Dev. 1996;10(10):1284–95. . [DOI] [PubMed] [Google Scholar]
- 53.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318(5858):1931–4. doi: 10.1126/science.1149460 [DOI] [PubMed] [Google Scholar]
- 54.Luo Y, Henricksen LA, Giuliano RE, Prifti L, Callahan LM, Federoff HJ. VIP is a transcriptional target of Nurr1 in dopaminergic cells. Experimental neurology. 2007;203(1):221–32. doi: 10.1016/j.expneurol.2006.08.005 [DOI] [PubMed] [Google Scholar]
- 55.Jackson JJ, Lindemann MD, Boling JA, Matthews JC. Summer-Long Grazing of High vs. Low Endophyte (Neotyphodium coenophialum)-Infected Tall Fescue by Growing Beef Steers Results in Distinct Temporal Blood Analyte Response Patterns, with Poor Correlation to Serum Prolactin Levels. Front Vet Sci. 2015;2:77 doi: 10.3389/fvets.2015.00077 ; PubMed Central PMCID: PMCPMC4685929. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Within a sequence, underlined nucleotides indicate the forward and reverse primer positions.
(DOCX)
The red and blue dots represent linear combinations of the relative expression data, including expression values and variances, of the 26,675 gene transcripts in each Bovine GeneChip.
(DOCX)
As indicated by the legend color box, white color in the middle represents the mean value, 0; red color represents gene expression levels above the mean expression; and blue color denotes expression below the mean. The intensity of the color reflects the relative intensity of the fold change.
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files, except for the .cel files which are available from GEO (http://www.ncbi.nlm.nih.gov/geo/) as accession no. GSE62570.