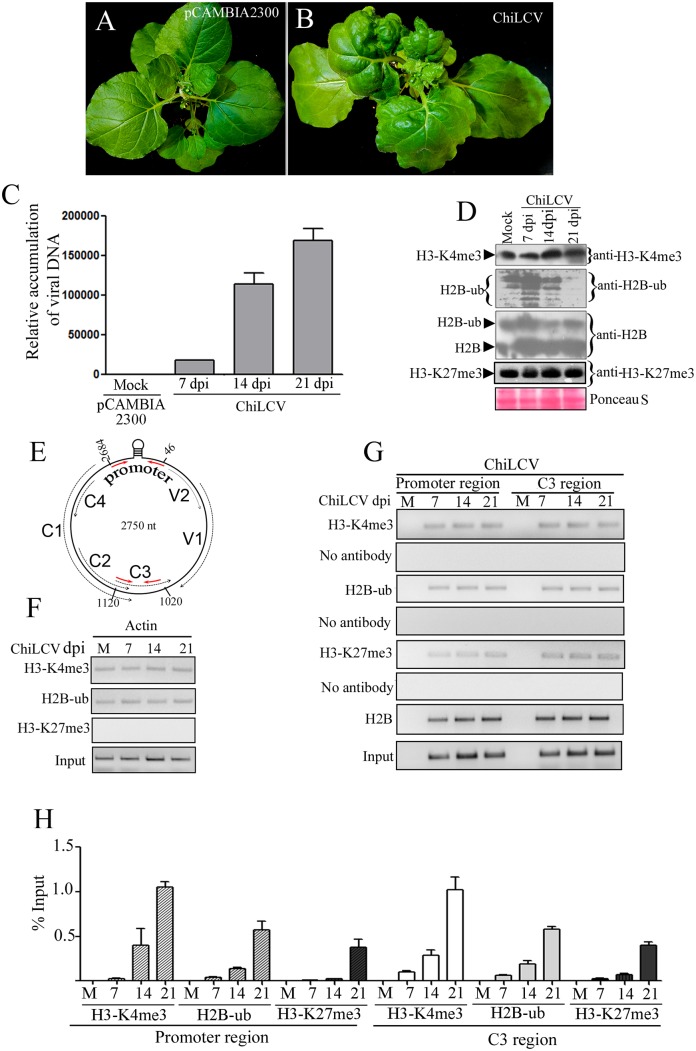
Fig 1. Deposition of H2B-ub and H3-K4me3 on the ChiLCV genome.
(A) An N. benthamiana plant inoculated with the vector pCAMBIA2300. (B) Phenotype of a representative N. benthamiana plant inoculated with ChiLCV showing typical symptoms of leaf curl disease at 21 days post inoculation (dpi). (C) qPCR of viral DNA accumulation at different dpi. (D) Immunoblot analysis of global cellular H2B, H2B-ub, H3-K4me3 and H3-K27me3 levels in mock-, and virus-inoculated N. benthamiana plants at 7, 14 and 21 dpi. Immunoblotting was performed using anti-H3-K4me3, anti-H2B-ub, anti-H2B and anti-H3-K27me3 specific antibodies following standard protocol. (E) Schematic diagram of the circular genome of ChiLCV (2750 nt) indicating the relative positions of the viral ORFs and the position of the primers (red arrows) used for chromatin immunoprecipitation in the study. (F) Detection of occupancy of H3-K4me3, H2B-ub and H3-K27me3 on the Actin genic region by ChIP-PCR serves as control. (G-H) Detection of H3-K4me3, H2B-ub, H3-K27me3 and H2B on the promoter and C3 region of ChiLCV by ChIP-PCR at different time points following infection using anti-H2B, anti-H2B-ub, anti-H3-K4me3 and H3-K27me3 antibodies and primers specific to the ChiLCV promoter (2684–46 nt) and the C3 region (1020–1120 nt).