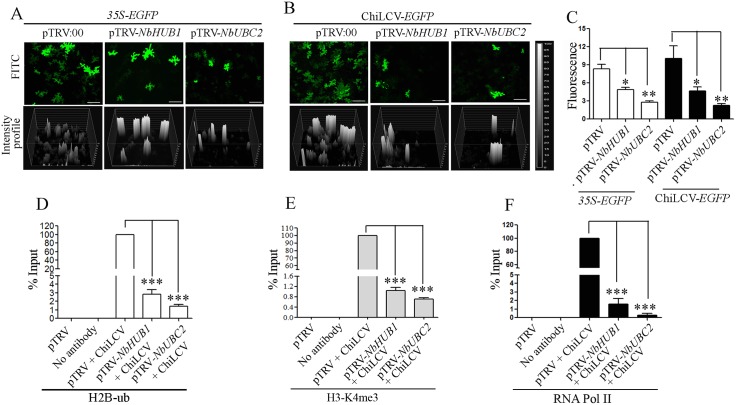
Fig 3. An analysis of comparative study of activity of ChiLCV and 35S promoter in silenced plants.
(A) 35S-EGFP constructs and (B) ChiLCV based EGFP expression vector (ChiLCV-EGFP). Scale bar = 100μm. Intensity profile of each samples was developed using NIS-Element 4.0 software. (C) Graphical representation of intensity of EGFP fluorescence of 35S-EGFP in pTRV:00 (n = 244), pTRV-NbHUB1 (n = 410) and pTRV-NbUBC2 (n = 385). EGFP intensity of ChiLCV-EGFP in pTRV:00 (n = 225), pTRV-NbHUB1 (360), pTRV-NbUBC2 (n = 410). n = total number of cells observed. (D-F) Detection of H2B-ub, H3-K4me3 and DNA- dependant RNA Polymerase II occupancy on ChiLCV promoter in NbUBC2-, and NbHUB1-silenced plants. ChIP-PCR was carried out using anti-H2B-ub, anti-H3-K4me3, RNA Pol II antibodies and ChiLCV promoter specific primers. The asterisk denotes statistical significance differences between mean values determined by t-test (***p<0.001, **p<0.01, *p<0.05).