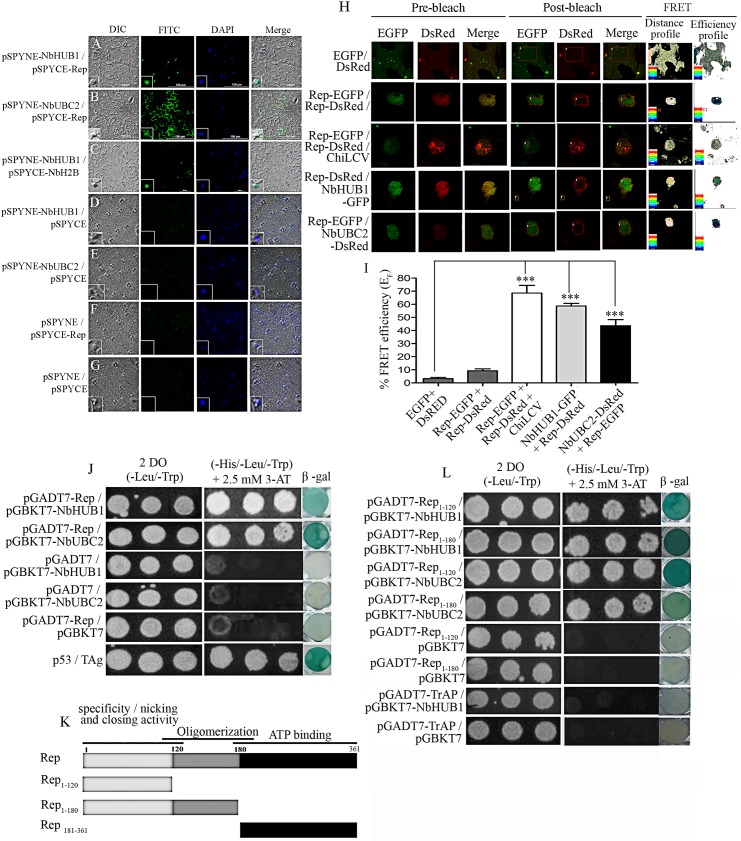
Fig 7. Rep interacts with NbHUB1 and NbUBC2 in vivo.
In planta bimolecular fluorescence complementation assays were performed in the lower epidermis of N. benthamiana leaves (at 5 dpi). NbHUB1 and NbUBC2 were expressed as in-frame fusion with the N-terminal of the YFP protein using the pSPYNE vector. The Rep protein was expressed with C-terminal region of the YFP protein using pSPYCE vector. BiFC assay of interaction of (A) pSPYNE-NbHUB1 and pSPYCE-Rep, (B) pSPYNE-NbUBC2 and pSPYCE-Rep, (C) pSPYNE-NbHUB1 and pSPYCE-NbH2B. (D) pSPYNE-NbHUB1 / pSPYCE, (E) pSPYNE-NbUBC2 / pSPYCE, (F) pSPYCE-Rep / pSPYNE and (G) pSPYNE / pSPYCE serve as control. Scale bar = 100μm. (H) The protein–protein interaction was also monitored by FRET microscopy in the epidermal cells of N. benthamiana leaves coexpressing EGFP and DsRed, Rep-EGFP and Rep-DsRed, NbHUB1-GFP and Rep-DsRed, Rep-EGFP and NbUBC2-DsRed. Representative acceptor photobleaching images show EGFP (donor) and DsRed (acceptor) channels before and after bleaching. After bleaching, DsRed fluorescence decreases in the bleached are as indicated by rectangles/circles. The FRET profiles showed the proximity between the donor and acceptor molecules and the efficiency of FRET. (I) Graph represents the quantification of FRET efficiency (EF) of three independent experiments. (J) Yeast two-hybrid assay of Rep and NbHUB1, Rep and NbUBC2 on non selective media (-Leu / -Trp) and selective media (-His / -Leu / -Trp with 2.5 mM 3-AT). β galactosidase activity were checked for each combination and corresponding negative controls.P53 and TAg served as positive control. (K) Schematic diagram of deletion mutants of Rep protein used in yeast two-hybrid assays. (L) Yeast two-hybrid and β-galactosidase assays of in vivo interaction between deletion mutants of Rep protein and TrAP with NbHUB1 and NbUBC2.