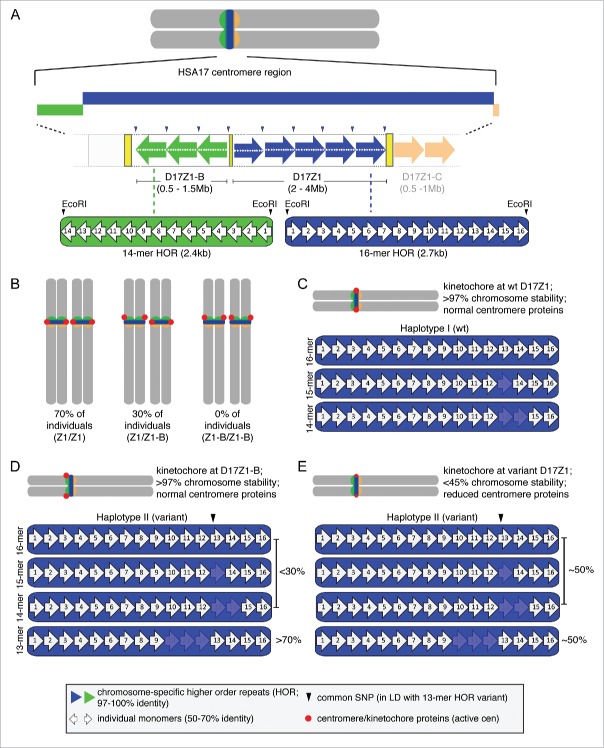
Figure 1.
α satellite variation and the molecular basis of centromeric epialleles in humans. (A) The centromere region of human chromosome 17 (HSA17) contains 3 α satellite arrays that are each defined by a different higher order repeat (HOR) unit. α satellite DNA is composed of 171bp monomers (white arrows) that are 50–70% identical. A defined number of monomers are tandemly arranged to create a HOR that is chromosome-specific. D17Z1 (blue), the predominant array is defined by a canonical 16-monomer HOR; EcoRI restriction sites demarcate the first monomer of each HOR. D17Z1-B (green) and D17Z1-C (shaded orange) are each defined by different 14-mer HORs. The monomers are numbered by their order in the HOR, and do not necessarily indicate sequence identity at the same monomer position between HORs of different arrays. (B) In the population, 70% of individuals carry 2 HSA17s that assemble the centromere and kinetochore (red dot) at D17Z1 (Z1/Z1). In 30% of individuals (Z1/Z1-B), D17Z1-B is the active centromere on one HSA17 homolog and D17Z1 is the centromere on the other homolog. No individuals have been identified yet that assemble both HSA17 centromeres at D17Z1-B (Z1-B/Z1-B). (C) D17Z1 is a polymorphic array. Single and multiple monomeric deletions produce HOR variants, including 15-mers, 14-mers, 13-mers, as well as 12-mers and 11-mers (not shown). (D) Some monomers also carry a common SNP in monomer 13 (black arrowhead) that creates an EcoRI site. This SNP is in linkage disequilibrium (LD) with the 13-mer HOR. Arrays containing specific HORs and the SNPs exist as distinct haplotypes in humans. Wild-type haplotype (I) occurs in most individuals and is defined by the canonical 16-mer HOR, as well as rarer 15- and 14-mers (C). Wild-type D17Z1 arrays are usually the site of centromere and kinetochore assembly (red circles) on mitotically stable HSA17. Haplotype II is defined by HOR variants that include a high proportion of 13-mers, many of which contain the SNP. D17Z1 arrays that have a high proportion of variant HORs are less likely to be the site of centromere assembly. Instead, the centromere is formed at “backup” array D17Z1-B and the HSA17 is extremely stable. (E) In a subset of Haplotype II individuals, the proportion of wild-type to variant HORs within the multi-megabase D17Z1 array is nearly equivalent. In these instances, if the centromere forms at D17Z1, the HSA17 is extremely unstable in mitosis due to a deficiency in centromere and kinetochore proteins (small red circles) and abnormal kinetochore architecture.