FIGURE 1:
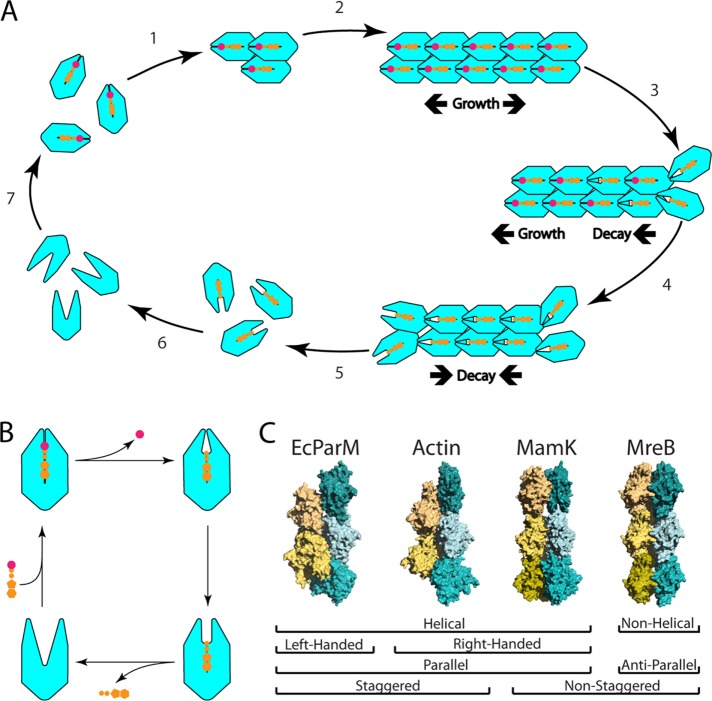
Polymer formation is a repeated feature within the actin superfamily. (A) Polymerization of actin homologues requires the formation of a filament nucleus (1). Once a nucleus has formed, filament elongation is rapid (2). Once monomers incorporate into a growing filament, they begin to hydrolyze ATP (3) until the filament end(s) is(are) composed of ADP-bound monomers (4). Filaments initially grow in both directions. Under certain conditions, some filaments can grow from one end and shrink from the other (a process referred to as treadmilling). Eventually, lower affinity between ADP-bound monomers allows for filament disassembly from the ends (5). ADP then dissociates from ADP-bound monomers (6), which then rapidly rebind ATP (7). (B) ATP hydrolysis, phosphate release, ADP/ATP exchange are associated with changes in monomer conformation that influence filament architecture and actin on/off rates. (C) Among actin-like filaments, the contacts within individual protofilaments vary little. By varying lateral contacts between protofilaments, different filament structures with different properties and behaviors can be generated. A subset of these structures is displayed here. E. coli ParM is a parallel, left-handed, helical filament whose protofilaments are staggered (PDB ID: 5AEY) (Bharat et al., 2015). Actin is a parallel, helical filament whose protofilaments are staggered (PDB ID: 4A7N) (Behrman et al., 2012). MamK filaments are parallel, helical filaments whose protofilaments are not staggered (PDB ID: 5LJV) (Löwe et al., 2016). MreB is a nonhelical, antiparallel filament whose protofilaments are not staggered and that has an intrinsic curvature (PDB ID: 4CZJ) (van den Ent et al., 2014).