FIGURE 1:
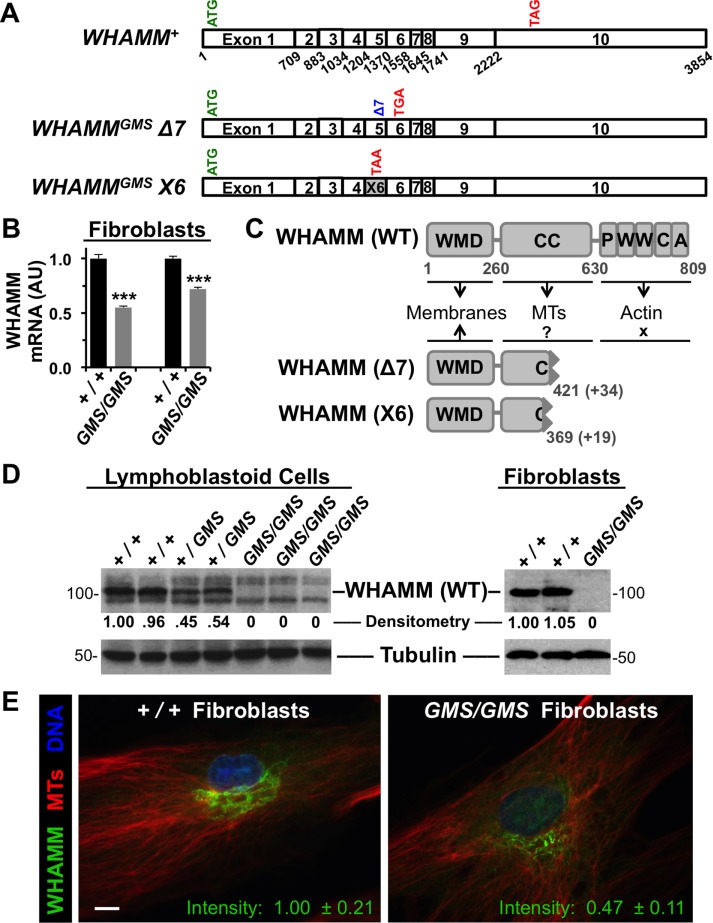
Cells from Amish GMS patients encode truncated WHAMM variants. (A) Diagrams of the exon organization in wild-type WHAMM cDNA and in the Δ7 and X6 variants are shown. Start and stop codons are indicated in green and red, respectively. (B) RNA was isolated from normal (+/+) or Amish GMS patient (GMS/GMS) fibroblasts and subjected to RT-PCR with primers to exons 9–10 of WHAMM or to β-actin. Relative levels of WHAMM-specific products (mean + SEM from three to four experiments) are shown. ***, p < 0.001 (t tests). (C) The 809-residue WHAMM(WT) protein includes a WMD that interacts with membranes, a CC region that binds microtubules (MTs), and a C-terminal PWWCA segment that promotes actin nucleation. The GMS Δ7 and X6 variants include the N-terminal 421 or 369 amino acids of WHAMM followed by 34 or 19 additional residues after the respective frameshifts. (D) Lymphoblastoid cell lines and skin fibroblasts from homozygous unaffected (+/+), heterozygous (+/GMS), or homozygous affected (GMS/GMS) individuals were subjected to immunoblotting with anti-WHAMM (left), anti-WWCA (right), and anti-tubulin antibodies. Relative levels of WHAMM(WT) were determined by densitometry of a representative blot. (E) Wild-type (+/+) or GMS patient (GMS/GMS) fibroblasts were stained with antibodies to visualize WHAMM (green) and microtubules (red), and with DAPI to label DNA (blue). The fluorescence intensity of WHAMM staining (mean ± SEM from 10–12 cells per genotype) was lower in patient cells, p < 0.001 (t test). Scale bar: 10 μm.