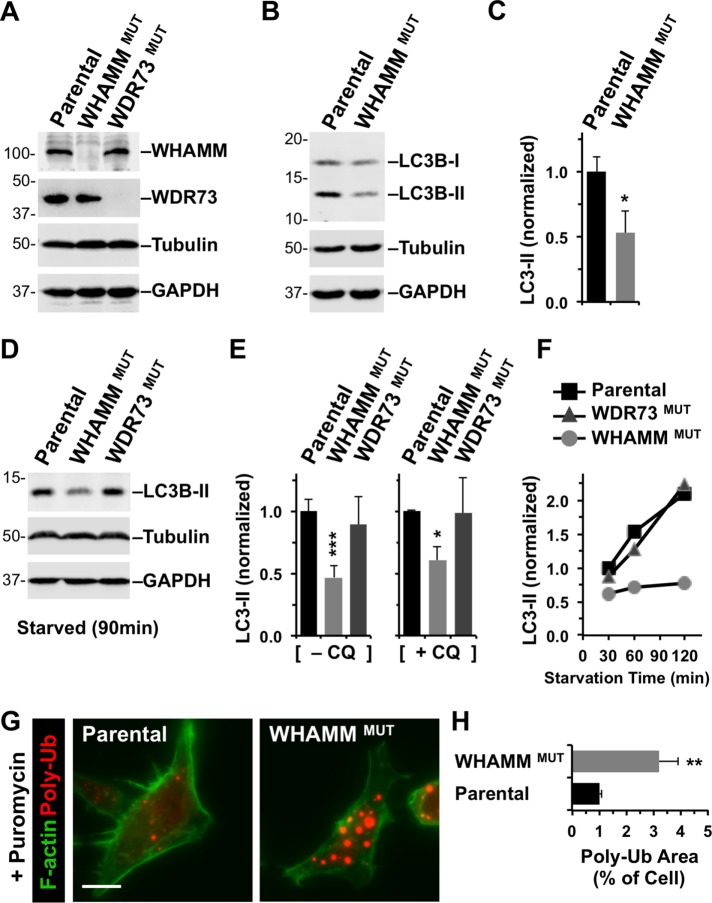
FIGURE 7:
Targeted inactivation of WHAMM inhibits lipidation of LC3 and clearance of protein aggregates. (A) Parental eHAP cells or cells with mutations engineered in WHAMM or WDR73 were analyzed by immunoblotting for WHAMM, WDR73, tubulin, and GAPDH. (B) eHAP and WHAMMMUT cells were analyzed by immunoblotting for LC3B, tubulin, and GAPDH. LC3B was detected in its immature (LC3-I) and mature lipidated (LC3-II) forms. (C) LC3B-II band intensities were quantified relative to tubulin. Each bar represents the mean (+ SD) from analyses of three experiments. *, p < 0.05 (t test). (D, E) eHAP cell derivatives were starved for 90 min in the absence or presence of chloroquine (CQ), and LC3-II levels were measured as in B and C. Each bar represents the mean (+ SD) from analyses of three to five experiments. *, p < 0.05; ***, p < 0.001 (ANOVA). (F) eHAP cell derivatives were starved for the indicated times, and LC3-II levels were measured as in C. (G) Parental or WHAMM-deficient cells were treated with puromycin for 3.5 h and stained with antibodies to visualize ubiquitinated protein aggregates (red) and with phalloidin to visualize F-actin (green). Scale bar: 10 μm. (H) The areas occupied by ubiquitinated protein aggregates and the entire cell were calculated using ImageJ and converted to a ratio. Each bar represents the mean (+ SEM) from analyses of 30–35 cells. **, p < 0.01 (t test).