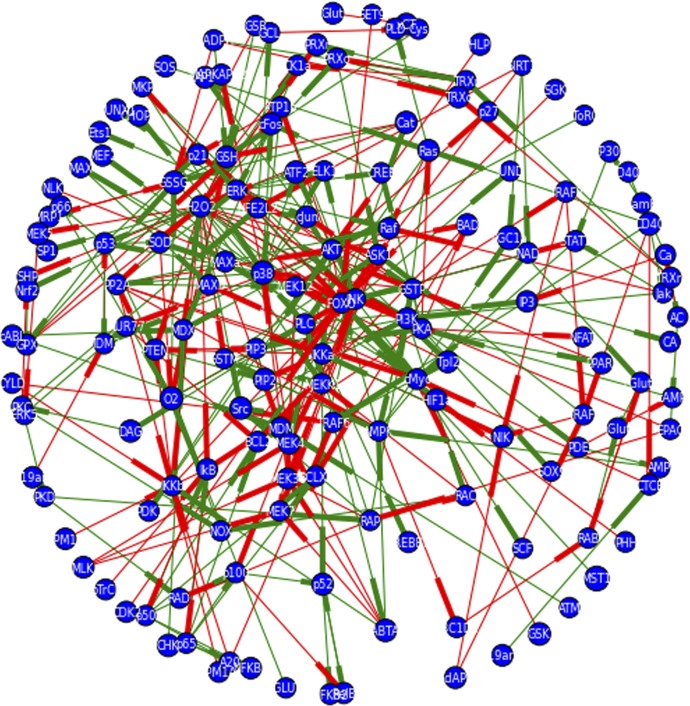
Figure 4. Network graph of signaling pathways induced by CD40L and glutamate and transcription factors, which may be activated by these pathways that are predicted to bind near the PSGL-1 start site.
Primary and secondary sources were consulted for signaling interactions that might mediate signaling downstream of CD40L, glutamate, or GSH depletion via manual curation of search results from PubMed. Additionally, signaling databases, such as KEGG pathways, were consulted. The ENCODE database was consulted to identify transcription factors that might bind around the PSGL-1 start site, and those for which a connection was found with the signaling pathways induced by CD40L or glutamate were encoded. All signaling interactions were coded as binary interactions in Python using NetworkX and matplotlib libraries. Negative regulatory interactions are signified by a red edge, whereas positive regulatory interactions are signified by a green edge. Directionality of the interaction is indicated by the weighting of the side of the edge closest to the regulated node. The network contains 155 nodes and 341 edges.