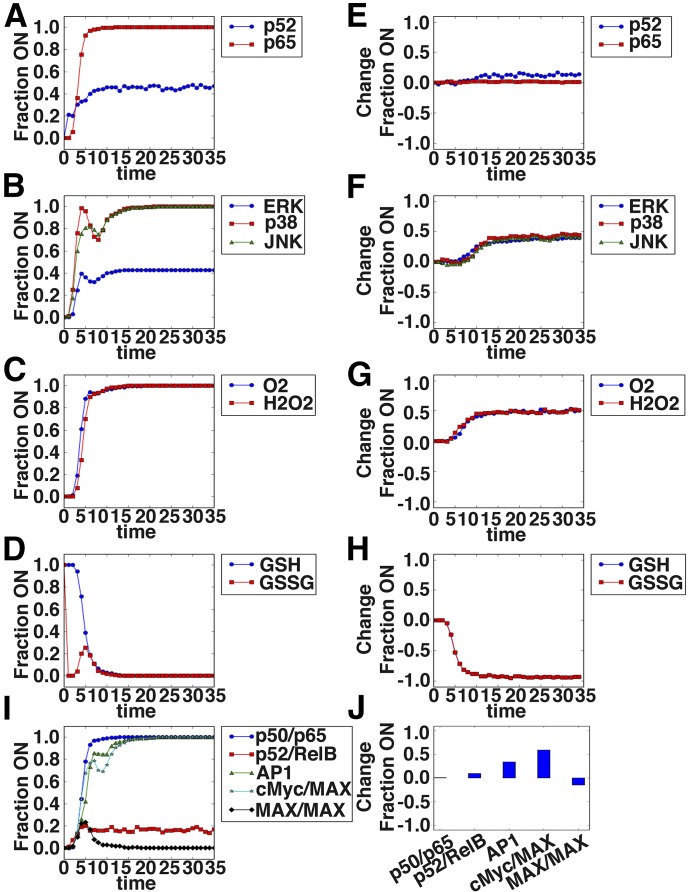
Figure 6. Boolean network model reveals signaling dynamics downstream of sCD40L + glutamate treatment of monocytes and predicts c-Myc as a likely regulator of PSGL-1 induction.
The network illustrated in Fig. 4 was implemented in Python using the Boolean net library, with log rules describing the regulatory interaction between nodes inferred based on the literature describing the regulation. A total of 800 simulations, with 80 time steps, of the CD40L + glutamate-alone scenario were run using an asynchronous update schedule. The fraction of simulations in which a node was on or had a value of 1 is shown for selected signaling intermediates in A–D. The difference in the percentage of simulations in which a node was “on” between the combination scenario and CD40L-alone scenario is shown in E–H. A positive value indicates increased activation in the combination scenario. (A and E) The NF-κB family members p52 and p65 are shown. (B and F) The MAPK family members ERK, p38, and JNK are shown. (C and G) The reactive oxygen intermediates O2 and H2O2 are shown. (D and H) The reduced GSH and oxidized GSSG form of GSH are shown. Shown are the dynamics of the transcription factor complexes p50/p65, p52/RelB, cFos/cJun (AP-1), c-Myc/MAX, and MAX/MAX (I) and the change in the steady-state values relative to CD40L alone (J).