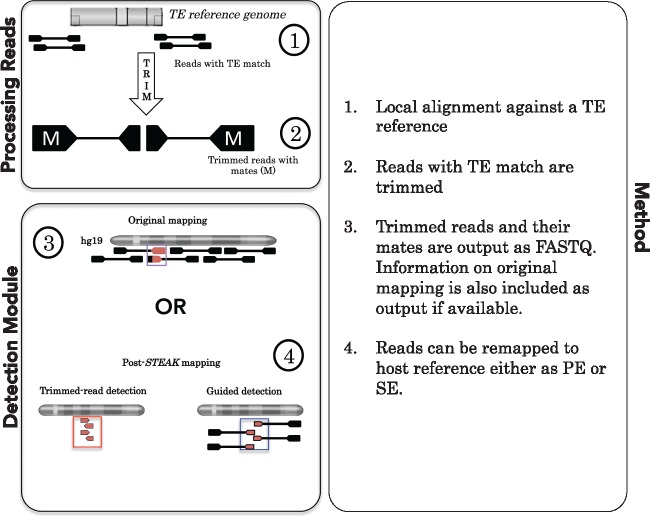
Figure 1.
Workflow of STEAK. Processing data: All reads are locally aligned using the Smith Waterman algorithm and allowing mismatches when mapping reads against a TE reference (5′- and 3′-ends and respective reverse complements). Reads that match with the TE are trimmed of the matching portion. Information on the trimmed reads and their mates, such as the original mapping positions, MAPQ, and sequence qualities, are kept in STEAK outputs. Detection Module: Trimmed reads can be remapped to the human reference either as single-end (trimmed read detection) or paired-end reads (guided detection).