Figure 9.
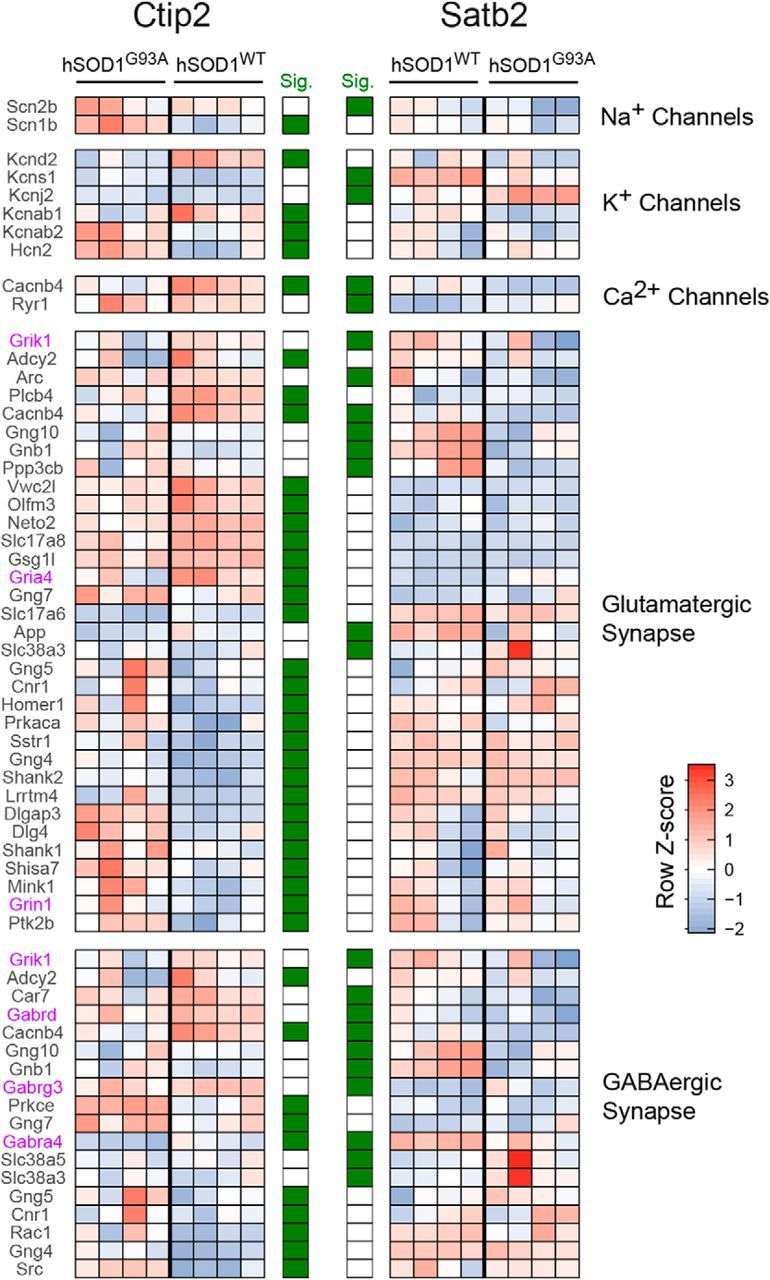
Differential modulation of genes for ion channels and synaptic proteins in corticospinal and corticocortical neurons in an ALS mouse model. Heatmap of significantly differentially expressed genes from select, physiologically relevant gene sets. Genes were selected as significant from differential expression analyses between hSOD1G93A and hSOD1WT in either CSN-enriched (CTIP2-positive) or CCN-enriched (SATB2-positive) cellular populations. Row z scores across all 16 samples are used to highlight the changes in expression profiles for each gene across the four conditions. Genes called significant using cuffdiff2 (5% FDR, Benjamini–Hochberg corrected) for CSN or CCN populations are marked in green in the significance columns. Genes for glutamate or GABA receptor subunits are shown in purple. Gene sets were hand curated from publicly available annotations including Reactome, MGI, and HUGO. Supporting data are found in Figure 8-1 available at https:/10.1523/JNEUROSCI.0811-17.2017.f8-1.
