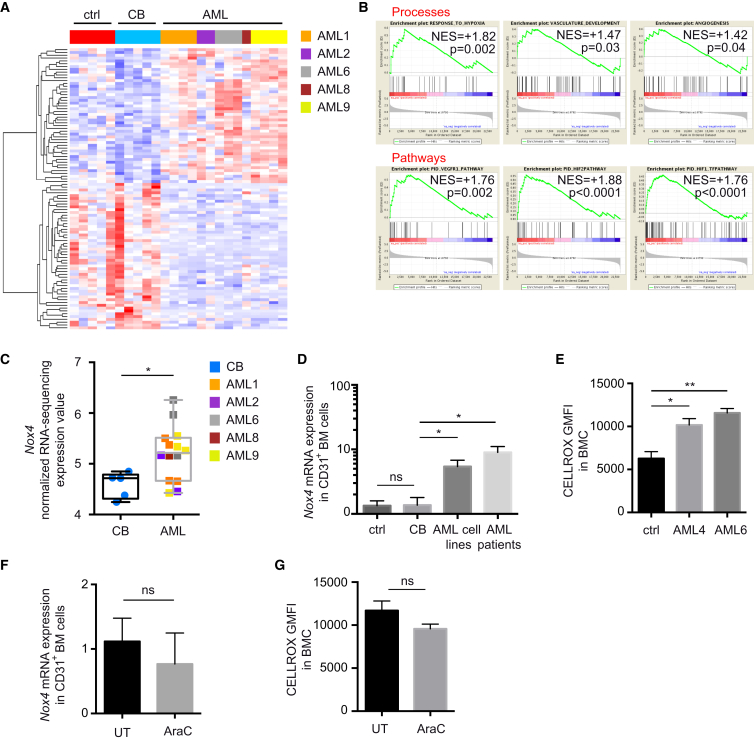
Figure 4.
AML-Induced Transcriptional Signature in BM ECs
(A) Heatmap from hierarchical clustering of GSEA top 100 ranked genes between untransplanted mice (ctrl group), mice engrafted with human CB-derived HSPCs (CB group), and mice engrafted with different patient-derived samples (AML group) derived BM ECs, using RNA-seq data. Red indicates higher expression values and blue indicates lower expression values (log2 FPKM [fragments per kilobase per million mapped] scale).
(B) Enrichment plots for Processes (top) and Pathways (bottom) from GSEA between CB and AML groups using RNA-seq gene expression data indicate enrichment of these pathways in AML group. Normalized enrichment score (NES) and nominal p value are shown.
(C) Normalized expression values of Nox4 in CB versus AML groups analyzed via RNA-seq. Data are shown as whiskers minimum-to-maximum plots, the line inside the box representing the mean, the top and the bottom lines representing the 75% and 25% percentiles, respectively, and the lines above and below the box representing the SD.
(D) Nox4 expression analyzed by qRT-PCR in CD31+ BM cells retrieved from the depicted groups of mice. CB, n = 5; AML cell lines, n = 8; AML patients (AML1, 2, 9), n = 13. Data are shown as mean ± SEM.
(E) Cellular ROS levels in BMC retrieved from mice of depicted groups; n = 3 per group. Data are shown as mean ± SEM.
(F) Nox4 expression analyzed by qRT-PCR in CD31+ BM cells retrieved from mice engrafted with AML6 and treated or not with AraC. n = 3 per group. Data are shown as mean ± SEM.
(G) Cellular ROS levels in BMC retrieved from mice engrafted with AML6 and treated or not with AraC; n = 3 per group. Data are shown as mean ± SEM.
ns, not significant; ∗p < 0.05, ∗∗p < 0.01. See also Figure S4.