Abstract
Interindividual variability in polymorphic uridine diphosphate-glucuronosyltransferase 1A1 (UGT1A1) ascribed to genetic diversity is associated with relative glucuronidation level among individuals. The present research was aimed to study the effect of 2 important single nucleotide polymorphisms (SNPs; rs8330 and rs10929303) of UGT1A1 gene on glucuronidation status of acetaminophen in healthy volunteers (n = 109). Among enrolled volunteers, 54.13% were male (n = 59) and 45.87% were female (n = 50). The in vivo activity of UGT1A1 was investigated by high-performance liquid chromatography-based analysis of glucuronidation status (ie, acetaminophen and acetaminophen glucuronide) in human volunteers after oral intake of a single dose (1000 mg) of acetaminophen. The TaqMan SNP genotyping assay was used for UGT1A1 genotyping. The wild-type genotype (C/C) was observed the most frequent one for both SNPs (rs8330 and rs10929303) and associated with fast glucuronidator phenotypes. The distribution of variant genotype (G/G) for SNP rs8330 was observed in 5% of male and 8% of the female population; however, for SNP rs10929303, the G/G genotype was found in 8% of both genders. A trimodal distribution (fast, intermediate, and slow) based on phenotypes was observed. Among the male participants, the glucuronidation phenotypes were observed as 7% slow, 37% intermediate, and 56% fast glucuronidators; however, these findings for the females were slightly different as 8%, 32%, and 60% respectively. The k-statistics revealed a compelling evidence for good concordance between phenotype and genotype with a k value of 1.00 for SNP rs8330 and 0.966 for SNP rs10929303 in our population.
Keywords: acetaminophen glucuronide, APAP, bilirubin, HPLC, Pakistan, paracetamol, UGT, UGT1A1
Introduction
The drug-metabolizing enzymes are responsible for biotransformation and final excretion of xenobiotics and drugs by increasing their hydrophilicity. Biotransformation of drugs involved the cytochrome P450-based phase I reactions (oxidation, reduction, and hydrolysis) and phase II reactions. In phase II of drug metabolism, there are conjugation actions like glucuronidation by uridine diphosphate (UDP) glucuronosyltransferases (UGTs), acetylation by N-acetyltransferase (NAT), and glutathione-S-transferase in the liver, with numerous proteins accountable for transportation. The enzymes involved in both phases are highly polymorphic. Interindividual variability exists in the comparative extent of phase I and II enzyme activities that influence biological responses to drug and xenobiotic exposure.1,2
The UGT (EC 2.4.1.17) is a microsomal enzyme existing in almost all living beings such as bacteria, animals, plants, and humans. The UGT is responsible for the glucuronidation of several exogenous compounds and drugs from almost all therapeutic classes and endogenous bilirubin. Glucuronidation accounts for 40% to 70% of xenobiotic elimination approximately. Through glucuronidation, small lipophilic molecules got conjugated with UDP and more water-soluble molecules are formed.3 The removal of a broad range of xenobiotics as a consequence of UGT-mediated conjugation explains the drug detoxifying and transformation potential of this enzyme.3,4
Acetaminophen (acetyl-para-aminophenol [APAP]/paracetamol) is an extensively used analgesic over-the-counter drug. The APAP biotransformation is based on its conjugation with glucuronic acid catalyzed by UGT. This conjugation reaction results in a more water-soluble APAP molecule by shifting the glucuronosyl moiety from UDP-glucuronic acid to acetaminophen. As the glucuronidation is a major metabolic pathway of APAP, the acetaminophen glucuronide is the primary drug metabolite that can be identified in the blood.5
The glucuronidation activity studied among different ethnic groups is well differentiated as a bimodal and a trimodal distribution. Based on the glucuronidation status, the individuals among a population may be categorized as fast and slow glucuronidators (bimodal model)6 or as fast, intermediate, and slow glucuronidators (trimodal model).7,8 The slow glucuronidation phenotype is widespread in Yorubans (50%), Caucasians (23%), and Japanese (16%). The fraction of fast and slow glucuronidators varies among different ethnic divisions.9
The superfamily UGT is divided into 4 subfamilies on account of amino acid sequence homology, named UDP-glucuronosyltransferase 1, 2, 3, and 4. The isozymes of family UGT1A have the first exon that is spliced into 2 to 5 common exons and thus producing a C and N-terminal domain. The gene-specific promoter region is possessed by each member of UGT1A family.10 In human, 13 isoenzymes from UGT1A gene, located on chromosome 2q37, are known to be originated as a consequence of a genetic alteration in exon 1 region; however, 6 isoforms of the UGTB2 subfamily are encoded by a rigid cluster of separate genes located on chromosome 4.11
Both UGT1A and UGT2B genes are highly polymorphic. Polymorphisms of UGT1A1 gene are known to be associated with variation in glucuronidation status and consequently with the susceptibility of a number of diseases. Commonly, a single nucleotide polymorphism (SNP) in UGT1A1 results in a reduced expression and enzyme activity of UGT that predispose to a clinical condition known as jaundice observed in Crigler-Najjar syndrome, type 1 and type 2.12 Interindividual variability in acetaminophen glucuronidation capacity is ascribed to the polymorphism of UGT1A1 gene. The polymorphism UGT1A1*28 is associated with reduced glucuronidation of bilirubin by hepatic UGTs, observed in Gilbert Syndrome and also results in altered acetaminophen metabolism.13,14
Genotypic and phenotypic characterization of UGT1A1 among healthy population may endorse the emerging concept of “Precision Medicine” based on the valuable information regarding genetic polymorphism and associated individuals’ capacity of glucuronidation. In the present study, the effect of UGT1A polymorphism (rs8330 and rs10929303) was studied on glucuronidation status of acetaminophen among local healthy male and female volunteers.
Materials and Methods
The study was approved by the institutional ethics committee and written informed consent was taken from each volunteer. The volunteers were asked to maintain an overnight fasting and then blood sample (3 mL through venepuncture) was collected before and after 1.5 hours of oral administration of a single dose of acetaminophen (Panadol, 1000 mg). The blood samples were processed for cell-free plasma (centrifuged at 5000 rpm for 10 minutes) and stored at −20°C till further analysis.
Study Population
A total number of 109 human volunteers (Pakistani nationals) were enrolled including 59 males (54.13%) and 50 females (45.87%), with an average age of around 22 years. The demographic data are given in Table 1. The baseline clinical examination and lab studies were performed to ensure the normal health status of all study participants. The volunteers were allowed to have a normal diet and alcohol, coffee and other caffeinated drinks were prohibited.
Table 1.
Comparative Demographic Data of Healthy Male and Female Participants.
| Parameters | Male (n = 59) | Female (n = 50) | |||||
|---|---|---|---|---|---|---|---|
| Mean (±) SD | Min | Max | Mean ± SD | Min | Max | ||
| Age (years) | 22.2 (2.4) | 19 | 32 | 22.3 (2.4) | 18 | 29 | |
| Weight (kg) | 69.8 (10.5) | 46 | 102 | 55.8 (9.4) | 40 | 80 | |
| Height (cm) | 170.9 (7.9) | 154.9 | 187.9 | 160.2 (4.4) | 144.8 | 170.2 | |
| Body temp (°F) | 98.2 (0.3) | 98 | 98.6 | 98.2 (0.4) | 98.2 | 98.6 | |
| Blood pressure (mm Hg) | Systolic | 116.8 (4.7) | 110 | 120 | 115.8 (5.8) | 100 | 130 |
| Diastolic | 77.5 (5.1) | 70 | 90 | 80 (7.3) | 70 | 90 | |
Abbreviations: Min, minimum; Max, maximum; SD, standard deviation; n, number of participants.
Exclusion Criteria
Individuals having a history of any allergic response to nonsteroidal anti-inflammatory drugs including acetaminophen and any clinical illness such as diabetes, hepatitis, hypertension, and/or pregnancy were excluded.
Chemicals and Reagents
The reference standard of acetaminophen (purity >99.0%) and acetaminophen β-d-glucuronide (analytical standard) were purchased from Sigma-Aldrich, St. Louis, Missouri, USA. Acetaminophen (Panadol) manufactured by Pharmatec Pvt Ltd (Pakistan) was purchased from the market. All the solvents and chemicals used were HPLC grade with high purity. Genomic DNA isolation kit was purchased from Biobasic Inc, Amherst, New York, USA and the RedSafe™ staining solution and TaqMan probes from Applied Biosystems, Foster City, California, USA. Deionized distilled water was obtained from Central High Tech Lab (Advanced GS-590, Distillery, and CPW-200 Japan), University of Agriculture, Faisalabad. Drug-free plasma was obtained from Chiniot Dialysis Centre, Faisalabad.
Determination of Acetaminophen Glucuronidation (Phenotype)
The UGT1A1 activity was evaluated with HPLC-based modified Vertzoni et al15 method measuring the glucuronidation status of the acetaminophen used as a probe drug.
Frozen blood specimen was thawed prior further processing for HPLC analysis for plasma acetaminophen and acetaminophen glucuronide. The deproteinization was achieved through mixing 100 µL plasma (of each specimen) with 200 µL of perchloric acid (15%) followed by vortex mixing for 1 minute and finally centrifuged at 13,000 rpm (16 110 × g) for 10 minutes. The supernatant was filtered through syringe filters (0.22 µm). The 20 µL of deproteinized ultrafiltrate was injected into HPLC column [Hypersil BDS-C18, 250 mm × 4.6 mm, internal diameter 5 µm (Thermo Electron Corporation, Waltham, MA, USA). The elution was achieved using a mobile phase constituted by aqueous buffer solution of 0.05 M KH2PO4 (475 mL), 5% acetic acid (pH 6.5), and methanol (25 mL) on isocratic mode at a flow rate of 1 mL/min.
The method employed was found to be highly reliable and sensitive and the lower limit of quantification for acetaminophen was 2.5 µg/mL and for acetaminophen glucuronide, it was 1 µg/mL. The calibration curve was constructed based on 6 standard concentrations, and a good linearity was observed at the concentration range of 2.5 to 200 µg/mL (R 2 ≥0.9949 for APAP and R 2 >0.9908 for APAP-Glu). The glucuronidation status was determined on the basis of the metabolic ratio of plasma acetaminophen glucuronide to acetaminophen (APAP-Glu/APAP), and cut-off values were identified through the probit plot.
Determination of UGT1A1 Polymorphism
Isolation of genomic DNA
The genomic DNA was isolated from venous blood previously stored at −20°C immediately after drawn from volunteers in EDTA anticoagulated tubes (BD Vacutainer®) using genomic DNA isolation kit (EZ-10 Spin Column for Genomic DNA Minipreps) manufactured by Bio-Basic Inc, Amherst, New York, USA. Isolated DNA was quantified on Gen5™, run on agarose gel electrophoresis (1% w/v) stained with RedSafe staining solution and documented under ultraviolet light in gel documentation system. High-quality DNA samples obtained were stored at −40°C for further analysis.
TaqMan SNP genotyping assay for UGT1A1 gene polymorphism
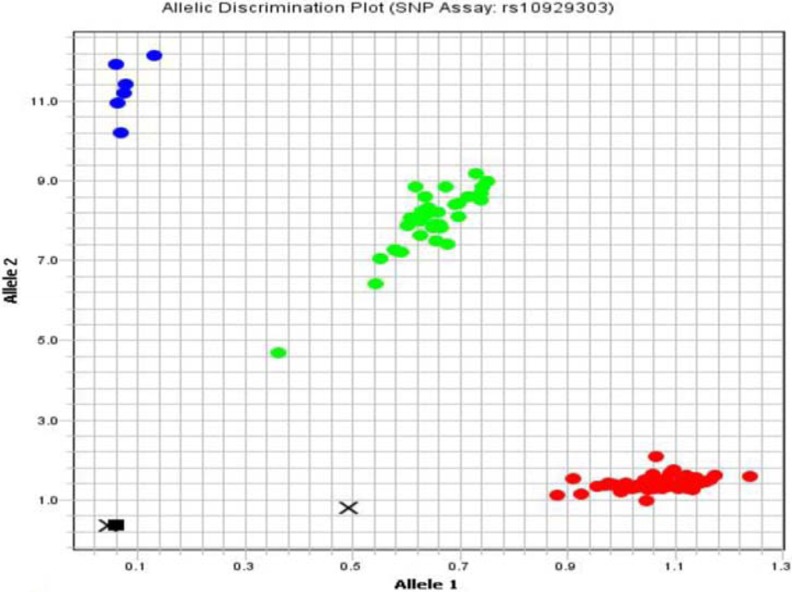
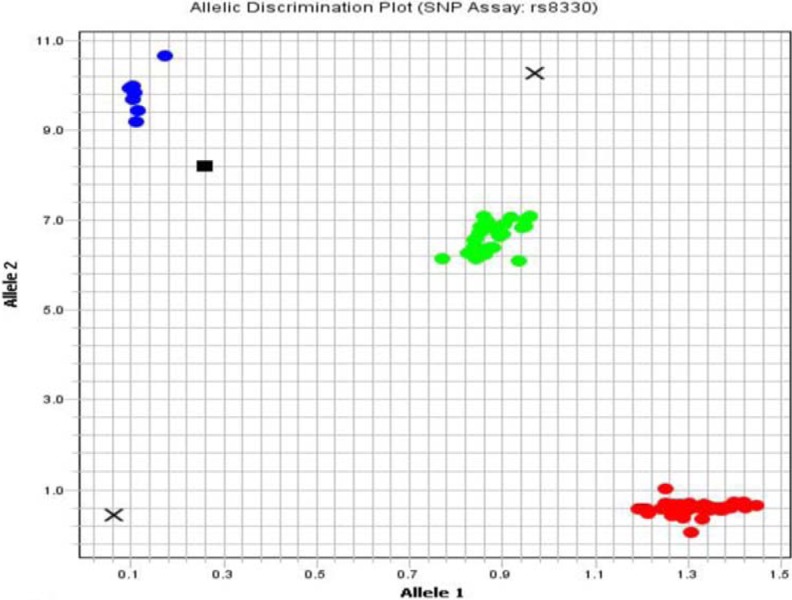
SNPs (rs10929303, c.1813C>T; rs8330, c.2042C>G) of the UGT1A1 gene in 3′ untranslated region (3′UTR) was studied by using sequence-specific TaqMan probes by real-time PCR (ViiA™ 7 System with Interchangeable block [96- and 384-well plate], OptiFlex™ System) manufactured by Applied Biosystem, USA. Oligonucleotide probes containing fluorescein amidite and VIC fluorescent dyes were used for UGT1A1 gene polymorphisms. Following the qPCR, the results were interpreted by allele discrimination plots (Figures 1 and 2) using ViiA 7™ Software, Applied Biosystem.
Figure 1.
Allele discrimination plot showing alleles as wild-type homozygous C/C (lower right cluster), mutant heterozygous allele C/T (middle cluster), and mutant homozygous T/T (upper right cluster) for the SNP rs10929303 by TaqMan SNP genotyping assay using ViiA 7™ Software, Applied Biosystem, USA. SNP indicates single nucleotide polymorphism.
Figure 2.
Allele discrimination plot showing alleles as wild-type homozygous C/C (lower right cluster), mutant heterozygous C/G (middle cluster), and mutant homozygous G/G (upper left) for the SNP rs8330 by TaqMan SNP genotyping assay using ViiA 7™ Software, Applied Biosystem, USA. SNP indicates single nucleotide polymorphism.
The DNA template (40 ng) was added in 384-well plate for drying prior to the addition of an optimized reaction mixture (5.0 µL) consisting of TaqMan genotyping master mix 2X (2.50 µL), TaqMan genotyping assay mix 20X (0.25 µL), and nucleases-free water (2.25 µL).
The PCR conditions optimized for qPCR consisted of a preread stage of 30 seconds at 60°C, a hold stage of 10 minutes at 95°C, PCR stage including denaturation at 95°C for 15 seconds, and annealing and amplification at 62°C and 72°C for 1 minute, respectively.
Statistical Analysis
The genotype and allelic frequencies were calculated using the Hardy-Weinberg equilibrium and presented in percentages. The frequency distribution histograms were formed using Minitab 15. The probit plot was used for the distribution of acetaminophen glucuronidation phenotype into fast, intermediate, and slow glucuronidators. K-statistics (κ test) was applied to measure the degree of agreement (concordance) between phenotype and genotype using SPSS Statistics (version 17). The comparison of parameters between male and female volunteers was evaluated by chi-square test.
Results
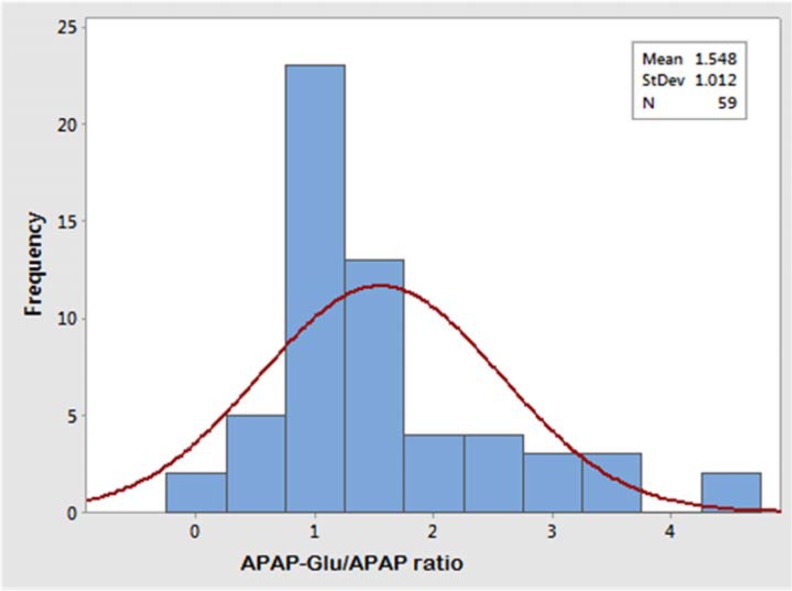
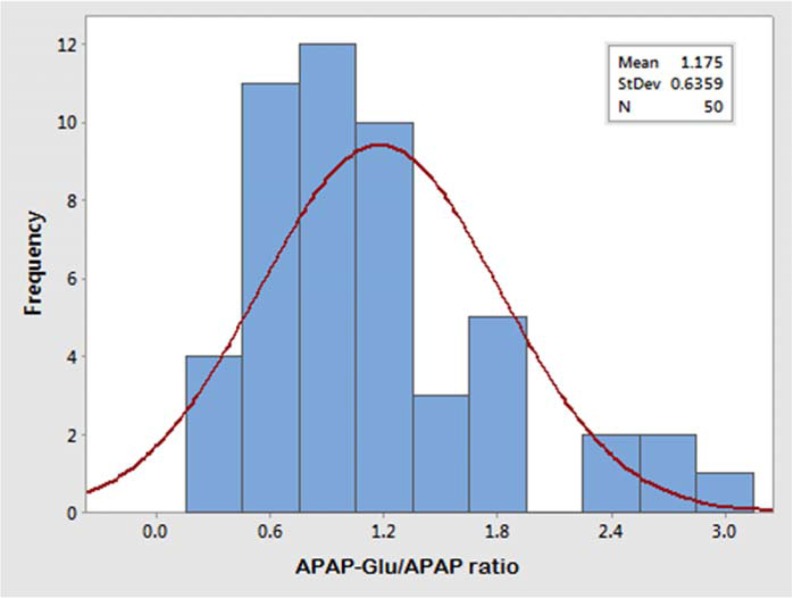
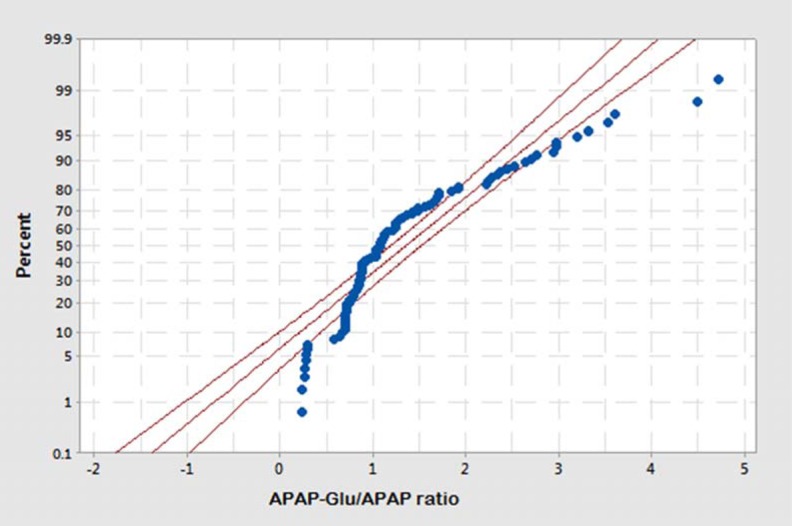
The glucuronidation status of UGT1A1 was determined in healthy male (n = 59) and female (n = 50) participants after a single oral dose of 1000 mg acetaminophen. The concentration of acetaminophen and acetaminophen glucuronide was measured in blood samples drawn at 1.5-hours following of its oral intake. The APAP-Glu/APAP metabolic ratio for the male volunteers ranged from 0.23 to 4.72, and 81% male volunteers have a metabolic ratio ranging from 0.2 to 2.8. The allocation of glucuronidation phenotype illustrated that 7% (n = 4) were slow, 37% (n = 22) were intermediate, and 56% (n = 33) male volunteers were fast glucuronidators phenotypically whereas in female population the APAP-Glu/APAP metabolic ratio ranged from 0.26 to 2.95, and 82% of female volunteers have metabolic ratio ranging from 0.6 to 1.8. The distribution of slow, intermediate, and fast glucuronidators among females was 8% (n = 4), 32% (n = 16), and 60% (n = 30), respectively. The frequency histograms of APAP-Glu/APAP metabolic ratio of male and female volunteers are shown in Figures 3 and 4, respectively. The volunteers who had a metabolic ratio less than or equal to 0.3 were found to be slow glucuronidators whereas those with a metabolic ratio of >0.3 were intermediate glucuronidators. The fast or rapid glucuronidators phenotypes were assigned to the volunteers with a metabolic ratio of ≥1.8. The cut-off points of metabolic ratios for discriminating the phenotypes were identified from probit plot as shown in Figure 5. The probit plot provides the information regarding how closely the data points follow the fitted distribution line. The distribution of glucuronidation phenotype was found a good fit, as the points fall closely along the straight lines. For a normal distribution achieved with APAP-Glu/APAP ratio, around 8% population has been observed to have slow glucuronidation phenotype.
Figure 3.
Frequency histogram of the metabolic ratio of APAP-Glu/APAP showing trimodal distribution (slow, intermediate, and fast) of healthy male volunteers (n = 59). APAP-Glu indicates acetyl-para-aminophenol-glucuronide.
Figure 4.
Frequency histogram of the metabolic ratio of APAP-Glu/APAP showing trimodal distribution (slow, intermediate, and fast) of healthy female volunteers (n = 50). APAP-Glu indicates acetyl-para-aminophenol-glucuronide.
Figure 5.
Probability plot of acetaminophen glucuronide/acetaminophen metabolic ratio in healthy male and female volunteers (n = 109) showing cut-off points for distribution of fast, intermediate, and slow glucuronidation phenotype.
A total of 109 human volunteers (male = 59, female = 50) were genotyped in this study. Two SNPs (rs10929303, rs8330) were studied in 3′ untranslated region of UGT1A1 gene. Mutant alleles for SNPs, rs10929303 and rs8330, were identified as alleles having a substitution at the positions of nucleotides, 1813 and 2042, respectively. In our study, the major genotypes for SNP (rs8330) are C/C & C/G and for SNP (rs10929303) are C/C & C/T, respectively, in both male and female. The frequency distribution of homozygous wild-type, heterozygous and homozygous mutant alleles, and the genotype frequency for both SNPs are summarized in Table 2.
Table 2.
Allele and Genotype Frequency Distribution of UGT1A1 (rs8330 and rs10929303) in Healthy Volunteers.
| UGT1A1 Polymorphisms | Genotypes | Genotype Frequency | Alleles | Allelic Frequency | ||
|---|---|---|---|---|---|---|
| Male (%) | Female (%) | Male (%) | Female (%) | |||
| rs8330 | C/C | 54.23 | 60 | C | 73.7 | 75.5 |
| C/G | 40.68 | 32 | ||||
| G/G | 5.08 | 8 | G | 26.2 | 24.4 | |
| rs10929303 | C/C | 52.54 | 62 | C | 72 | 75.5 |
| C/T | 38.98 | 30 | ||||
| T/T | 8.47 | 8 | T | 27.9 | 24.4 | |
The allelic frequencies of both SNPs (rs8330 and rs10929303) in male and female are in Hardy-Weinberg equilibrium. The genotype frequencies of UGT1A1 gene SNPs (rs8330 and rs10929303) differ insignificantly (p-value > 0.05) among male and female volunteers.
The concordance was calculated between UGT1A1 genotype and phenotype statistically. The k-statistics revealed an outstanding degree of association between phenotype and genotype with a κ value of 1.00 in both male and female participants for SNP rs8330 as shown in Table 3. The κ value also indicated good concordance between genotype (rs10929303) and phenotype (κ = 0.966) as shown in Table 4. No association of specific glucuronidator phenotypes was found between male and female volunteers (χ2 = 0.582; P = 0.748).
Table 3.
Comparison of Genotype (rs8330) and Acetaminophen Phenotype in Male and Female Volunteers.a
| Acetaminophen Phenotype | Genotype (rs8330) | Total | ||
|---|---|---|---|---|
| C/C | C/G | G/G | ||
| Fast | 62 | 0 | 0 | 62 |
| Intermediate | 0 | 39 | 0 | 39 |
| Slow | 0 | 0 | 8 | 8 |
| Total | 62 | 39 | 8 | 109 |
aκ value = 1.
Table 4.
Comparison of Genotype (rs10929303) and Acetaminophen Phenotype in Male and Female Volunteers.a
| Acetaminophen Phenotype | Genotype (rs10929303) | Total | ||
|---|---|---|---|---|
| C/C | C/T | T/T | ||
| Fast | 61 | 1 | 0 | 62 |
| Intermediate | 1 | 38 | 0 | 39 |
| Slow | 0 | 0 | 8 | 8 |
| Total | 62 | 39 | 8 | 109 |
aκ value = 0.969.
For SNP rs8330, the frequency distribution data of slow and intermediate glucuronidator phenotypes and the associated genotypes for male and female volunteers are given in Tables 5 and 6, respectively. The comparative data of genotypes for SNP rs10929303 and associated glucuronidator phenotypes in male and female volunteers are shown in Tables 7 and 8, respectively.
Table 5.
Comparison of Acetaminophen Phenotype/UGT1A1 (rs8330) Genotype in Male Volunteers.a,b
| Acetaminophen Phenotype (Male) | Genotype rs8330 (Male) | Total | ||
|---|---|---|---|---|
| C/C | C/G | G/G | ||
| Fast | 32 | 0 | 0 | 32 |
| Intermediate | 0 | 23 | 0 | 23 |
| Slow | 0 | 0 | 4 | 4 |
| Total | 32 | 23 | 4 | 59 |
an = 59.
bκ value = 1.
Table 6.
Comparison of Acetaminophen Phenotype/UGT1A1 rs8330 Genotype in Female Volunteers.a,b
| Acetaminophen Phenotype (Female) | Genotype rs8330 (Female) | Total | ||
|---|---|---|---|---|
| C/C | C/G | G/G | ||
| Fast | 30 | 0 | 0 | 30 |
| Intermediate | 0 | 16 | 0 | 16 |
| Slow | 0 | 0 | 4 | 4 |
| Total | 30 | 16 | 4 | 50 |
an = 50.
bκ value = 1.
Table 7.
Comparison of Acetaminophen Phenotype/UGT1A1 (rs10929303) Genotype in Male Volunteers.a
| Acetaminophen Phenotype (Male) | Genotype rs10929303 (Male) | Total | ||
|---|---|---|---|---|
| C/C | C/T | T/T | ||
| Fast | 31 | 1 | 0 | 32 |
| Intermediate | 0 | 23 | 0 | 23 |
| Slow | 0 | 0 | 4 | 4 |
| Total | 31 | 24 | 4 | 59 |
aκ value = 0.969.
Table 8.
Comparison of Acetaminophen Phenotype/UGT1A1 (rs10929303) Genotype in Female Volunteers.a,b
| Acetaminophen Phenotype (Female) | Genotype rs10929303 (Female) | Total | ||
|---|---|---|---|---|
| C/C | C/T | T/T | ||
| Fast | 30 | 1 | 0 | 31 |
| Intermediate | 0 | 15 | 0 | 15 |
| Slow | 0 | 0 | 4 | 4 |
| Total | 30 | 16 | 4 | 50 |
an = 50.
bκ value = 1.
Discussion
Interindividual differences in activity of drug metabolizing enzymes have been associated with the personalized responsiveness to the toxicants and drugs in a population. Glucuronidation is the best launched example of polymorphism in the drug metabolizing enzyme.16
A pathologically reduced UGT enzyme activity has been ascribed to the genetic polymorphism of UGT1A gene family, attributed to an altered transcriptional pattern and function of the resultant protein. The mutant alleles of UGT1A1 gene result in declined bilirubin conjugation capacity of UGT1A1 isoform. The UGT1A gene polymorphism also contributes to the individual’s susceptibility to various diseases owing to the altered process of inactivation and detoxification of xenobiotics and carcinogens.17
Kadakol et al18 summarized the data of approximately 50 polymorphisms of UGT1A1 gene contributing to Crigler-Najjar syndrome type I and II. The type I syndrome is a clinical condition presented with nonhemolytic jaundice, characterized by marked unconjugated hyperbilirubinemia and frequent central nervous system involvement because of no UGT1A1 activity. Another condition associated with UGT1A1 enzyme inactivity is Gilbert syndrome associated with a polymorphism in the promoter region of UGT1A1 gene.19 The differential expression and function of the UGT1A isoforms in the colon,20 liver,21 pancreas,22 and kidney cancers23 have been reported.
A higher risk of having colorectal carcinoma is reported among individuals carrying mutant alleles of UGT1A gene.24 The breast cancer susceptibility is also found to be associated with mutant however, no relationship has been observed yet between estrogen receptor status and the UGT1A1 genotypes.25,26
Acetaminophen glucuronide is a major conjugate formed during acetaminophen biotransformation among conjugates of sulfate, cysteine, and mercapturate as reported by Pabba et al27 in healthy Indian population.
In our research, the genetic polymorphism of UGT1A (rs8330 and rs10929303) in local population has been studied and reported for the first time in Pakistan. No authentic study has been accessed through standard databases and search engines with the focus of UGT1A1 genotypes being reported here (rs8330 and rs10929303) and their effect on the phenotypic distribution for Pakistani population.
The major aim of the research was to study the association or concordance between UGT1A1 genotype and glucuronidation phenotype. Our results showed a good association between UGT1A1 genotype for both SNPs (rs8330 and rs10929303) and capacity of acetaminophen glucuronidation in healthy volunteers.
The HPLC-based phenotypic studies for acetaminophen glucuronidation have been conducted in Hong Kong, Chinese, and Caucasian healthy population using acetaminophen as probe drug.28,29
Based on the individuals’ glucuronidation ability, accessed by plasma PAP-Glu/PAP ratio, the studied populations have presented a trimodal phenotypic distribution and the volunteers have been classified as fast, intermediate, and slow glucuronidators. The similar approach of phenotypic characterization of UGT1A1 through the ratio of morphine-glucuronide/morphine ratio, using morphine as probe drug, was reported by Holthe et al.8
In the present study, a comparable distribution of slow glucuronidator phenotype was observed between healthy male (7%) and female (8%) volunteers. The phenotypic distribution of intermediate and fast glucuronidators slightly differ among male and female volunteers. The volunteers with intermediate glucuronidator phenotype were 37% among males and 32% among females. There is a slightly higher percentage of female fast glucuronidators, that is, 60% versus 56% of male volunteers. However, on the basis of urine and saliva samples from healthy volunteers, a higher glucuronidation status have been reported among male participants as compared to the females.30 These noncomparable results of glucuronidation might be associated with the relative bioavailability, excretion, and elimination of acetaminophen in blood, saliva, and urine respectively.
Moreover, the divergent research findings among populations may be attributed to the ethnic differences owing to the particular genetic make-up of different populations. Besides pharmacogenetics, the geographical basis, environmental aspects, and diet (ie, differences in protein intake) may also contribute to the ethnic diversity of acetaminophen biotransformation.31 Distribution of UGT1A1 alleles (rs8330 and rs10929303) in our data shows resemblance with the Caucasian, Mexican, and Tuscan population.9
Conclusively, through our study, the UGT1A1 phenotypes encoded by wild-type alleles (C/C) for both SNPs (rs8330 and rs10929303) are categorized as fast glucuronidator based on their plasma acetaminophen glucuronidation status. Participants with mutant homozygous alleles for both SNPs (G/G, T/T, for rs8330 and rs10929303, respectively) were found to have slow glucuronidator phenotype. However, the individuals with heterozygous alleles were categorized as intermediate glucuronidators.
Acknowledgments
Authors highly acknowledge the Higher Education Commission, Pakistan, for providing funds for the research. For research facilities, authors duly appreciate the contribution of Department of Medicinal Chemistry and Molecular Pharmacology, College of Pharmacy, Purdue University West Lafayette, Indiana, USA, and Pharmaceutical Research Lab, Department of Biochemistry, University of Agriculture Faisalabad-Pakistan.
Authors’ Note: Huma Mehboob carried out the HPLC analysis of acetaminophen, acetaminophen glucuronide, DNA Isolation, Quantification, and RT-PCR of UGT1A1 SNPs (rs8330 and rs10929303). Huma Mehboob and Imtiaz Mahmood Tahir inferred the data and drafted the manuscript. All authors critically reviewed the manuscript before submission. Dr Huma Mehboob is the guarantor and principal investigator.
Declaration of Conflicting Interests: The author(s) declared no potential conflict of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: The research project was funded by Higher Education Commission, Islamabad, Pakistan.
References
- 1. Koo SH, Lee EJD. Pharmacogenetics approach to therapeutics. Clin Exp Pharmacol Physiol. 2006;33(5-6):525–532. [DOI] [PubMed] [Google Scholar]
- 2. Tahir IM, Iqbal T, Saleem S, Mehboob H, Akhter N, Riaz M. Effect of acetaminophen on sulfamethazine acetylation in male volunteers. Int J Immunopathol Pharmacol. 2016;29(1):17–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ahn SJ, Dermauw W, Wybouw N, et al. Bacterial origin of a diverse family of UDP-glycosyltransferase genes in the Tetranychus urticae genome. Insect Biochem Mol Biol. 2014;50:43–57. [DOI] [PubMed] [Google Scholar]
- 4. Desai AA, Innocenti F, Ratain MJ. UGT pharmacogenomics. Pharmacogenetics. 2003;13(8):517–523. [DOI] [PubMed] [Google Scholar]
- 5. Court MH, Freytsis M, Wang X, et al. The UDP-glucuronosyltransferase (UGT) 1A polymorphism c.2042C>G (rs8330) is associated with increased human liver acetaminophen glucuronidation, increased UGT1A exon 5a/5b splice variant mRNA ratio, and decreased risk of unintentional acetaminophen-induced acute liver failure. J Pharmacol Exp Ther. 2013;345(2):297–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jakobsson J, Ekström L, Inotsume N, et al. Large differences in testosterone excretion in Korean and Swedish men are strongly associated with a UDP-glucuronosyl transferase 2B17 polymorphism. J Clin Endocrinol Metab. 2006;91(2):687–693. [DOI] [PubMed] [Google Scholar]
- 7. Bock KW, Schrenk D, Forster A, et al. The influence of environmental and genetic factors on CYP2D6, CYP1A2 and UDP-glucuronosyltransferases in man using sparteine, caffeine, and paracetamol as probes. Pharmacogenetics. 1994;4(4):209–218. [DOI] [PubMed] [Google Scholar]
- 8. Holthe M, Rakvåg TN, Klepstad P, et al. Sequence variations in the UDP-glucuronosyltransferase 2B7 (UGT2B7) gene: identification of 10 novel single nucleotide polymorphisms (SNPs) and analysis of their relevance to morphine glucuronidation in cancer patients. Pharmacogenomics J. 2003;3(1):17–26. [DOI] [PubMed] [Google Scholar]
- 9. Thorisson GA. The international hapmap project web site. Genome Res. 2005;15(11):1592–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mackenzie PI, Walter Bock K, Burchell B, et al. Nomenclature update for the mammalian UDP glycosyltransferase (UGT) gene superfamily. Pharmacogenet Genomics. 2005;15(10):677–685. [DOI] [PubMed] [Google Scholar]
- 11. Turgeon D. Relative enzymatic activity, protein stability, and tissue distribution of human steroid-metabolizing UGT2B subfamily members. Endocrinology. 2001;142(2):778–787. [DOI] [PubMed] [Google Scholar]
- 12. Liu X, Cheng D, Kuang Q, et al. Association of UGT1A1* 28 polymorphisms with irinotecan-induced toxicities in colorectal cancer: a meta-analysis in Caucasians. Pharmacogenomics J. 2014;14(2):120–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Burchell B. Genetic variation of human UDP-glucuronosyltransferase. Am J Pharmacogenomics. 2003;3(1):37–52. [DOI] [PubMed] [Google Scholar]
- 14. Esteban A, Pérez-Mateo M. Heterogeneity of paracetamol metabolism in Gilbert’s syndrome. Eur J Drug Metab Pharmacokinet. 1999;24(1):9–13. [DOI] [PubMed] [Google Scholar]
- 15. Vertzoni MV, Archontaki HA, Galanopoulou P. Development and optimization of a reversed-phase high-performance liquid chromatographic method for the determination of acetaminophen and its major metabolites in rabbit plasma and urine after a toxic dose. J Pharm Biomed Anal. 2003;32(3):487–493. [DOI] [PubMed] [Google Scholar]
- 16. Cascorbi I. Genetic basis of toxic reactions to drugs and chemicals. Toxicol Lett. 2006;162(1):16–28. [DOI] [PubMed] [Google Scholar]
- 17. Guillemette C. Pharmacogenomics of human UDP-glucuronosyltransferase enzymes. Pharmacogenomics J. 2003;3(3):136–158. [DOI] [PubMed] [Google Scholar]
- 18. Kadakol A, Ghosh SS, Sappal BS, et al. Genetic lesions of bilirubin uridine-diphosphoglucuronate glucuronosyltransferase (UGT1A1) causing Crigler-Najjar and Gilbert syndromes: correlation of genotype to phenotype. Hum Mutat. 2000;16(4):297–306. [DOI] [PubMed] [Google Scholar]
- 19. Monaghan G, Ryan M, Hume R, et al. Genetic variation in bilirubin UDP-glucuronosyltransferase gene promoter and Gilbert’s syndrome. Lancet. 1996;347(9001):578–581. [DOI] [PubMed] [Google Scholar]
- 20. Bigler J, Whitton J, Lampe JW, et al. CYP2C9 and UGT1A6 genotypes modulate the protective effect of aspirin on colon adenoma risk. Cancer Res. 2001;61(9):3566–3569. [PubMed] [Google Scholar]
- 21. Tseng CS, Tang KS, Lo HW, et al. UDP-glucuronosyltransferase 1A7 genetic polymorphisms are associated with hepatocellular carcinoma risk and onset age. Am J Gastroenterol. 2005;100(8):1758. [DOI] [PubMed] [Google Scholar]
- 22. Piepoli A. Lack of association between UGT1A7, UGT1A9, ARP, SPINK1andCFTRgene polymorphisms and pancreatic cancer in Italian patients. World J Gastroenterol. 2006;12(39):6343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rouleau M, Roberge J, Bellemare J, et al. Dual roles for splice variants of the glucuronidation pathway as regulators of cellular metabolism. Mol Pharmacol. 2014;85(1):29–36. [DOI] [PubMed] [Google Scholar]
- 24. Tang KS. Link between colorectal cancer and polymorphisms in the uridine-diphosphoglucuronosyltransferase 1A7 and 1A1 genes. World J Gastroenterol. 2005;11(21):3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Guillemette C, Millikan RC, Newman B, et al. Genetic polymorphisms in uridine diphospho-glucuronosyltransferase 1A1 and association with breast cancer among African Americans. Cancer Res. 2000;60(4):950–956. [PubMed] [Google Scholar]
- 26. Adegoke OJ, Shu XO, Gao YT, et al. Genetic polymorphisms in uridine diphospho-glucuronosyltransferase 1A1 (UGT1A1) and risk of breast cancer. Breast Cancer Res Treat. 2004;85(3):239–245. [DOI] [PubMed] [Google Scholar]
- 27. Pabba S, Bolla S, Kandhagatla R, et al. Paracetamol metabolism in Indian population. Drug Res. 2011;52(10):769–772. [DOI] [PubMed] [Google Scholar]
- 28. Critchley J, Critchley L, Anderson P, et al. Differences in the single-oral-dose pharmacokinetics and urinary excretion of paracetamol and its conjugates between Hong Kong Chinese and Caucasian subjects. J Clin Phar Ther. 2005;30(2):179–184. [DOI] [PubMed] [Google Scholar]
- 29. Flouvat B, Leneveu A, Fitoussi S, et al. Bioequivalence study comparing a new paracetamol solution for injection and propacetamol after single intravenous infusion in healthy subjects. Int J Clin Pharmacol Ther. 2004;42(1):50–57. [DOI] [PubMed] [Google Scholar]
- 30. Navarro SL, Chen Y, Li L, et al. UGT1A6 and UGT2B15 polymorphisms and acetaminophen conjugation in response to a randomized, controlled diet of select fruits and vegetables. Drug Metab Disposition. 2011;39(9):1650–1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Critchley JA, Nimmo GR, Gregson CA, et al. Inter-subject and ethnic differences in paracetamol metabolism. Br J Clin Pharmacol. 1986;22(6):649–657. [DOI] [PMC free article] [PubMed] [Google Scholar]