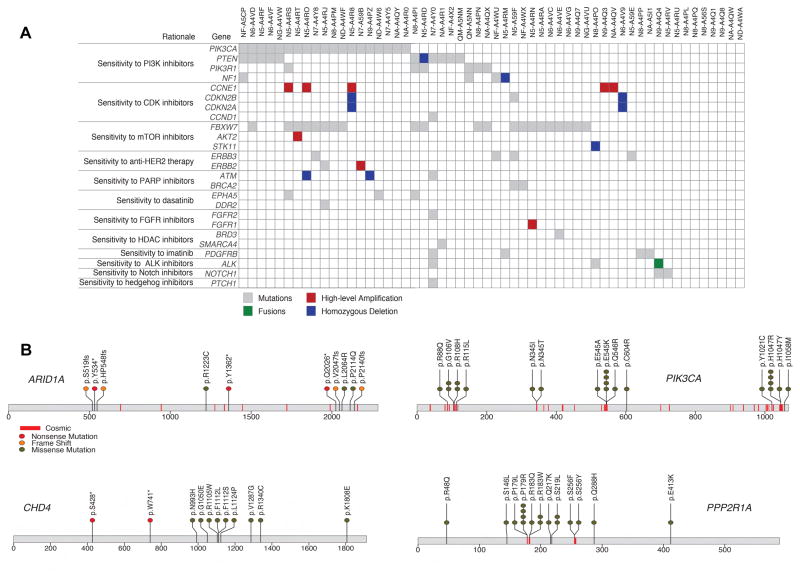
Figure 2. Clinically relevant alterations in uterine carcinosarcoma.
(A) Landscape of potentially clinically relevant mutations, copy number alterations and fusions in 56 UCS tumors, as identified by the PHIAL algorithm. Rows represent alterations in genes that lead to a given clinical rationale. Somatic mutations are shown in gray, homozygous deletions in blue, high-level amplifications in red (focal and more than two copies gained) and gene fusions in green. One POLE hypermutator case is excluded. (B) Mutation positions in amino acid coordinates for 4 of 14 significantly mutated genes. Each mutation is annotated by protein change (Gencode v18 transcripts). Also marked in red lines are sites of mutations from COSMIC v67 for which more than 5 experiments or samples had a mutation at the site.