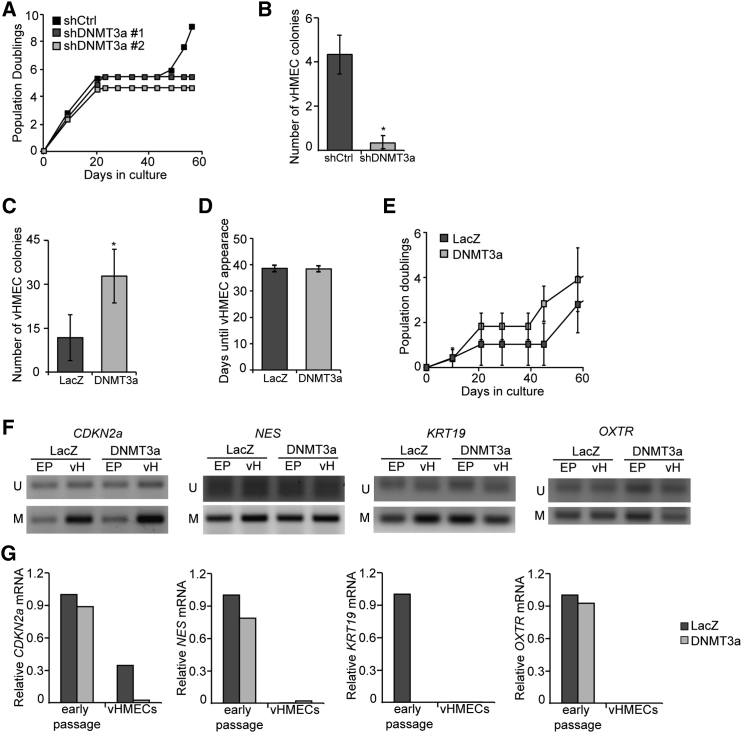
Figure 3.
vHMEC Reprogramming Is Partially Controlled by DNMT3a
(A) Growth curve showing cumulative population doublings over time in cultures with stable depletion of DNMT3a (shDNMT3a) by two different lentiviral short hairpins versus shCtrl.
(B) Quantitative analysis of the number of vHMEC colonies formed by cultures lacking DNMT3a (shDNMT3a) versus the firefly luciferase control (shCtrl), n = 3.
(C) Quantitative analysis of the number of vHMEC colonies formed by DNMT3a-overexpressing cultures versus the LacZ control, n = 4.
(D) Latency of vHMEC colony formation, defined as the number of days required for vHMECs to appear, in LacZ versus DNMT3a overexpressing cultures, n = 4.
(E) Growth curve showing cumulative population doublings over time in cultures with LacZ or DNMT3a overexpression, n = 3.
(F) Methylation-specific PCR of gene promoters in early-passage HMECs (EP) versus vHMECs (vH) expressing LacZ or DNMT3a.
(G) Representative image of qPCR analysis of expression of DNA methylation-regulated genes in early HMECs versus vHMECs expressing LacZ or DNMT3a.
In all panels, error bars indicate the mean ± SEM and replicates are individual patient samples. ∗p < 0.05, Student's t test.