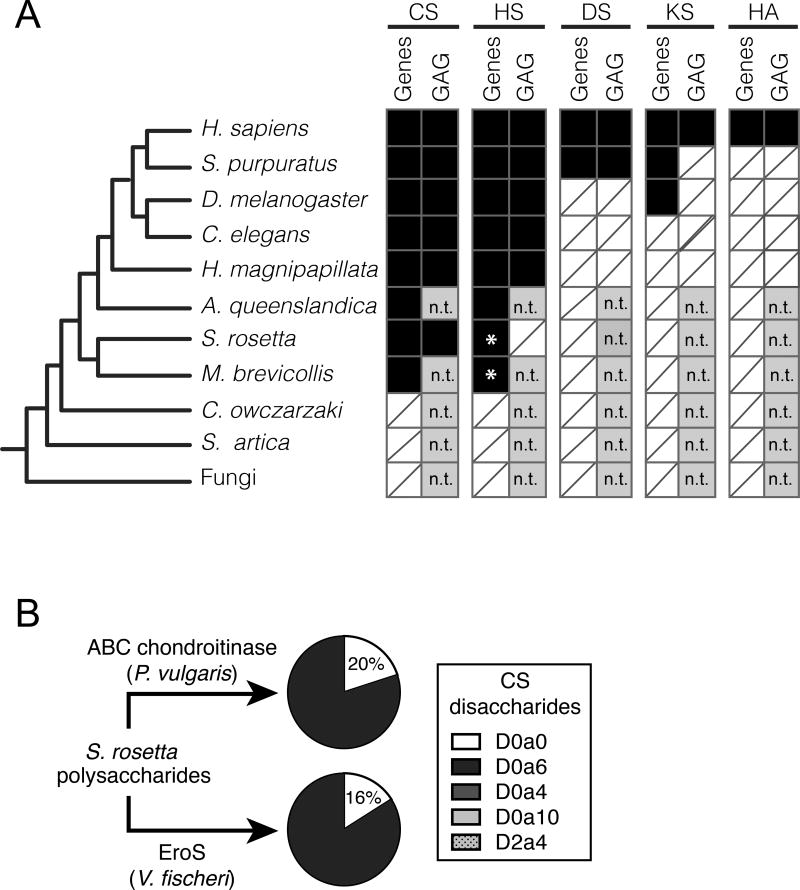
Figure 4. S. rosetta produces chondroitin sulfate that can be degraded by EroS.
(A) Phylogenetic distribution of diverse GAGs [CS= chondroitin sulfate; HS= heparan sulfate; DS= dermatan sulfate; KS= keratan sulfate; HA= hyaluronan], and their biosynthetic genes. The presence (black box) and absence (white box with slash) of genes required for the biosynthesis of GAGs (“Gene”) and biochemical evidence for GAGs (“GAG”) in S. rosetta and select opisthokonts. *; Ori et al. (2011) identified putative homologs of a subset of HS biosynthetic enzymes in the M. brevicollis genome, and we detect homologs of the same limited set of HS biosynthetic enzymes in S. rosetta. Importantly, these enzymes are shared components of the chondroitin biosynthetic pathway, and digestion of S. rosetta polysaccharides with heparinases failed to liberate heparan sulfate disaccharides, suggesting S. rosetta does not produce heparan sulfate (also refer to Figure S3). n.t.; not tested (experiments have not been performed to biochemically profile GAGs). (B) S. rosetta produces chondroitin that can be degraded by EroS. Polysaccharides isolated from S. rosetta were treated with either P. vulgaris ABC chondroitinase, an enzyme that can degrade many modifications of chondroitin into its disaccharide units (CS disaccharides), or purified EroS. Both ABC chondroitinase and EroS yielded similar amounts of unsulfated chondroitin disaccharide (D0a0) and chondroitin-6-sulfate disaccharide (D0a6) degradation products, indicating that unsulfated chondroitin and chondroitin-6-sulfate are produced by S. rosetta. In contrast, we were unable to detect chondroitin-4-sulfate (D0a4), chondroitin-4,6-sulfate (D0a10), or chondroitin-2,4-sulfate (D2a4) following degradation of S. rosetta polysaccharides with either EroS or ABC chondroitinase.