Figure 1.
Transcriptome Alterations in H1 hESC-Derived Lineages
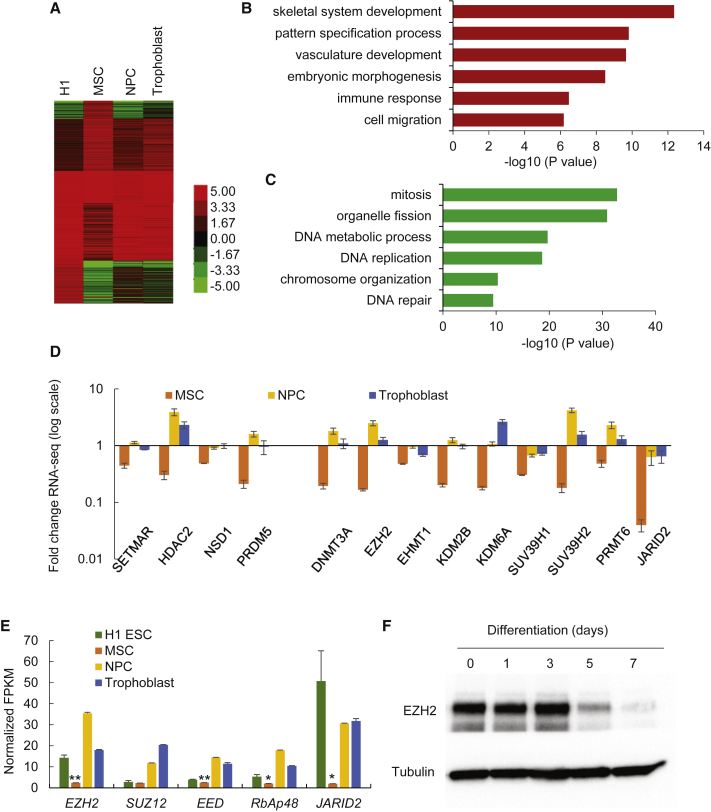
(A) Heatmaps showing the expression levels of coding genes in H1 hESCs and H1 hESC-derived cell lineages.
(B and C) Gene ontology (GO) enrichment analysis for upregulated genes (B) and downregulated genes (C) in H1 hESC-derived MSCs. The GO terms include biological function. Bars represent −log10 of p values.
(D) Fold changes of FPKM (fragments per kilobase of transcript per million mapped reads) value for genes encoding epigenetic modifiers in H1 hESC-derived lineages comparing with H1 hESCs. The y axis shows the log scale. Data are shown as mean ± SD from 2 replicates of RNA-seq data.
(E) Average FPKM value of genes encoding core components of PRC2 complex in H1 hESCs and H1 hESC-derived lineages. Data are shown as mean ± SD from 2 replicates of RNA-seq data. ∗p < 0.05; ∗∗p < 0.01.
(F) Western blot analysis of EZH2 level during differentiation of H1 hESCs to MSC.
See also Figure S1.