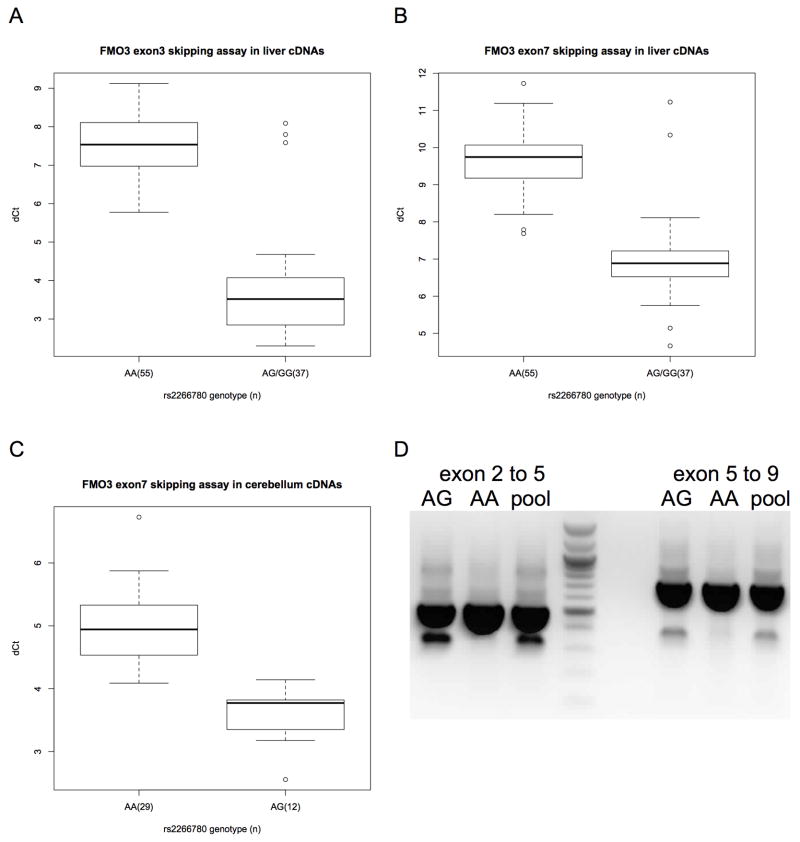
Figure 2.
Aberrant FMO3 splicing in human liver and cerebellum tissues by rs2266780 genotype. (A) Aberrant exon 2–4 splicing (skipping exon 3) versus correct exon 3–4 splicing in liver cDNAs, and (B) aberrant exon 6–8 splicing (skipping exon 7) versus correct exon 7–8 splicing in liver or (C) cerebellum cDNAs. The difference in PCR cycle times (ΔCt) for cDNAs for (n) samples divided by rs2266780 genotype. Relative expression, ΔCt, determined by subtracting the Ct value of the reaction using exon skipping primers from the Ct value of the reaction using correct splicing primers (see methods). The boxplot provides a summary of the data distribution. The box represents the interquartile range, which includes 50% of values. The line across the box indicates the median. The whisker lines extend to the highest and lowest values that are within 1.5x the interquartile range. Further outliers are marked with circles. (D) The products of PCR primers[39] flanking regions of FMO3 including exons 3 or 7 amplified from liver cDNAs heterozygous for s2266780 (AG), homozygous for the s2266780 major allele (AA) or from pooled cDNAs from eight liver samples of various genotypes.