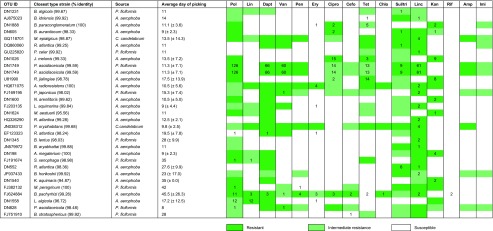
Table 2.
Resistance profiles of bacteria isolated from the sponges A. aerophoba, P. ficiformis and C. candelabrum
Bacterial identification was done by blastn of the 16S rRNA gene sequence (>800 bp) against a database of bacterial type strains (sequence identity values are shown). Numerical values indicate the number of times bacteria from the same OTU were detected on media supplemented with these antibiotics in the cultivation experiment from which they were derived. Note that no media were supplemented with ampicillin nor imipenem in the preceding cultivation experiment