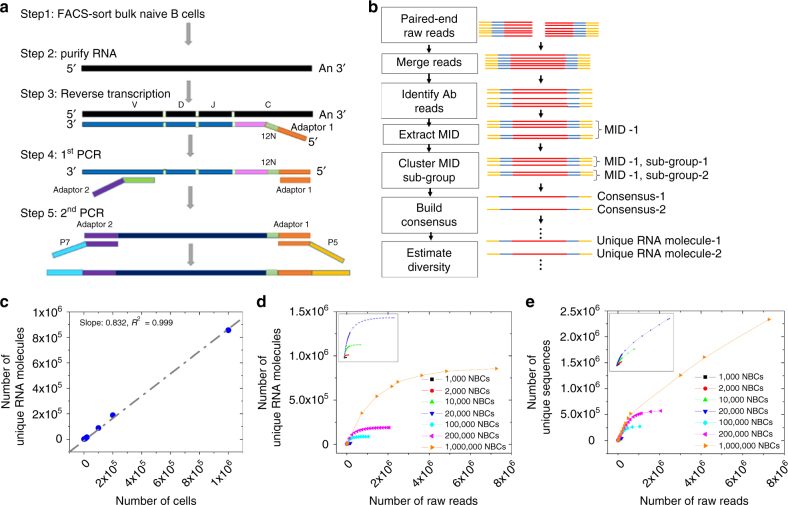
Fig. 1.
Ultra-accurate high coverage of antibody repertoire with a large dynamic range of input cells for MIDCIRS. a Schematic overview of tagging single immunoglobulin transcripts with MIDs. b Schematic overview of the informatics pipeline of MIDCIRS which includes merging paired-end reads, performing clustering to generate MID sub-groups, and building consensus sequences. c Correlation between number of cells and number of unique RNA molecules after using MIDCIRS. RNA from as few as 1000 to as many as 1,000,000 naive B cells was used as input material in generating the amplicon libraries. Slope indicates the estimated diversity coverage. d, e Rarefaction analysis of optimum sequencing depth for each sample with (d) and without (e) using MIDCIRS. NBCs naive B cells