Figure 1.
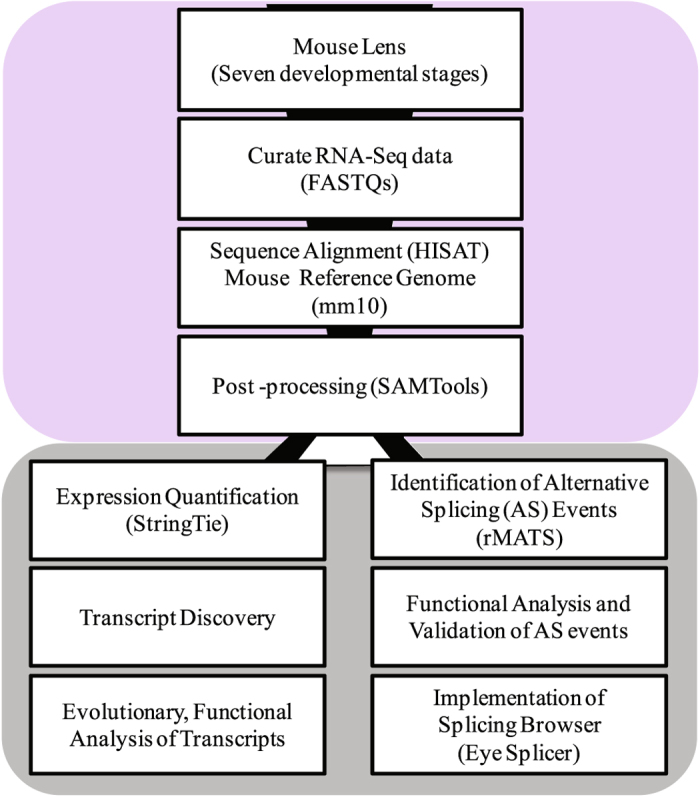
(a) Overview of the transcriptome analysis across developmental stages in mouse lens. Transcriptomes of mouse lens spanning seven developmental stages (three embryonic; E15, E15.5, E18 and four postnatal; P0, P3, P6, P9 stages with biological replicates) were collected from published sources for our study. Curated RNA sequence data was quality filtered using FASTX toolkit. High quality raw sequence reads were processed and aligned to mouse reference genome mm10 using HISAT and output collected as SAM files. Post processing (i.e. conversion of SAM to sorted BAM) of aligned reads was accomplished using SAMTools. Aligned and post processed RNA-Seq bam files associated with each developmental stage were utilized for two purposes. Firstly, for identifying and quantifying the expression levels of known and novel transcripts across seven developmental stages using StringTie, followed by an evolutionary and functional analysis to uncover high confident completely novel transcripts in developing lens. Secondly, the processed bam files were also employed for the identification of alternative splicing events using rMATS (replicate Multivariate Analysis of Transcript Splicing)27 followed by functional analysis of genes belonging to the enriched splice events. Finally, the results of the most prominent splicing events namely skipped exon and retained intron events are also made available through Eye splicer, a web based splicing browser showing developmentally altered splicing events in mouse lens.
