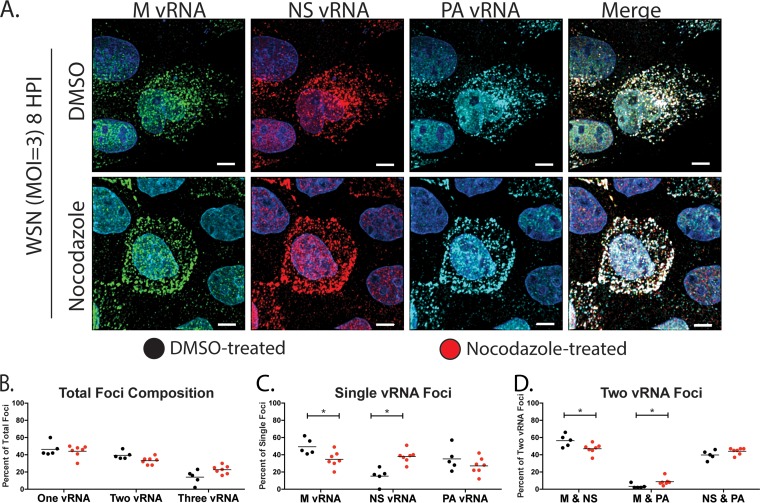
FIG 5.
Visualization of multiple vRNA segments in MDCK cells treated with nocodazole. (A) MDCK cells were infected with A/WSN/1933 (H1N1) and treated with DMSO or nocodazole (5 μg/ml) at 4 hpi and fixed at 8 hpi for FISH staining. FISH probes targeting the M vRNA segment (Alexa 488, green), NS vRNA (Quasar 570, red), and PA vRNA (Quasar 670, teal) were used. DAPI (blue) marks the cell nuclei, and the scale bars are 5 μm. Single representative cells chosen from the entire coverslip are displayed for clarity. (B to D) Finely spaced confocal stacks were acquired to reconstruct a 3D cell volume. Deconvolved stacks were analyzed via Imaris software to generate spots corresponding to the individual vRNA segments and to analyze the colocalization of spots. The proportions of foci containing one, two, or all three of the labeled vRNA segments are depicted in panel B. We further present a breakdown of the composition of each focus containing only one (C) or two (D) labeled vRNA segments. Each data point represents an individual cell that was analyzed by this method. A nonparametric Mann-Whitney test was performed to compare foci from DMSO (n = 5, black circles)- and nocodazole (n = 7, red circles)-treated cells.