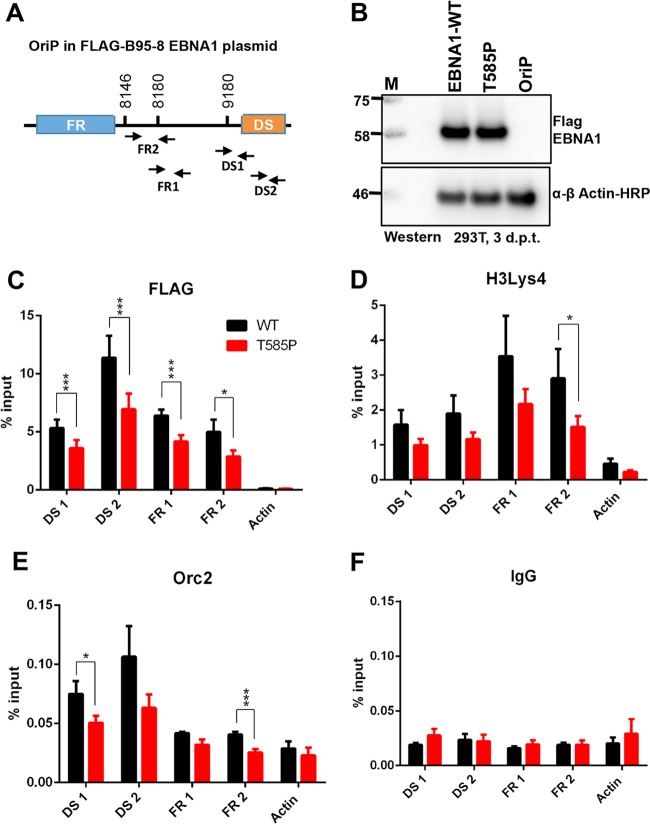
FIG 8.
The T585P mutation compromises EBNA1 chromatin binding and origin formation in vivo. (A) Schematic of the EBV OriP region in the plasmid and positions of primer sets used for ChIP-quantitative PCR. Numbers indicate B95-8 coordinates for primers. (B) Western blotting of 293T cells used for ChIP at 3 days posttransfection shows similar levels of WT EBNA1 and the T585P mutant. A vector with OriP lacking EBNA1 is used as a control to show the specificity of the Flag antibody for Flag-EBNA1. (C to F) Flag-EBNA1 T585P or Flag-WT EBNA1 was assayed by ChIP for Flag-EBNA1 (C), H3K4me3 (D), ORC2 (E), or control IgG (F) at DS and FR regions or for cellular actin as a control. The bar graphs represent the mean percentages of the input, and error bars represent standard errors of the means for three independent biological replicates, and within each replicate, PCR was done in duplicate. A GraphPad Prism t test was used to calculate P values (*, P < 0.05; ***, P < 0.001).