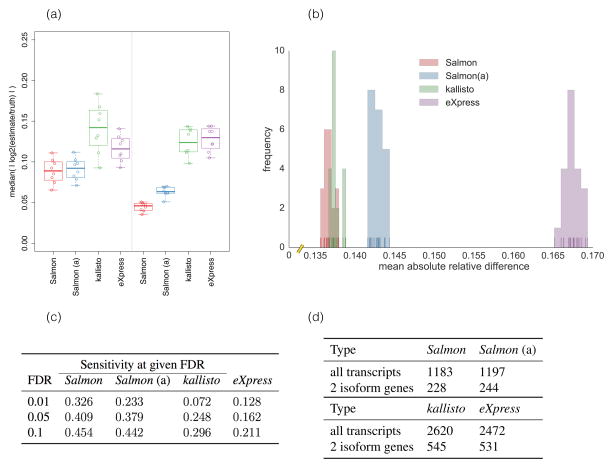
Figure 1.
(a) The median of absolute log fold changes (lfc) between the estimated and true abundances under all 16 replicates of the Polyester simulated data. The closer the lfc to 0, the more similar the true and estimated abundances. The left and right panels show the distribution of the log fold changes under samples simulated with different GC-bias curves learned from experimental data (details in Online methods, Ground truth simulated data). (b) The distribution of mean absolute relative differences (MARDs), as described in Online methods, Metrics for accuracy, of Salmon, Salmon using traditional alignments (“Salmon (a)”), kallisto and eXpress under 20 simulated replicates generated by RSEM-sim. Salmon and kallisto yield similar MARDs, though Salmon’s distribution of MARDs is significantly smaller (Mann-Whitney U test, p = 0.00017) than those of kallisto. Both methods outperform eXpress (Mann-Whitney U test, p = 3.39781 × 10−8). (c) At typical FDR values, the sensitivity of finding truly DE transcripts using Salmon’s estimates is 53%–450% greater than that using kallisto’s estimates and 210%–250% greater than that using eXpress’ estimates for the Polyester simulated data. (d) For 30 GEUVADIS samples, the number of transcripts called as DE at an expected FDR of 1% when the contrast between groups is simply a technical confound (i.e. the center at which they were sequenced). Salmon produces fewer than half as many DE calls as the other methods. Permuting samples, or testing for DE within sequencing center resulted in ≪ 1 transcript called as DE on average for all methods.