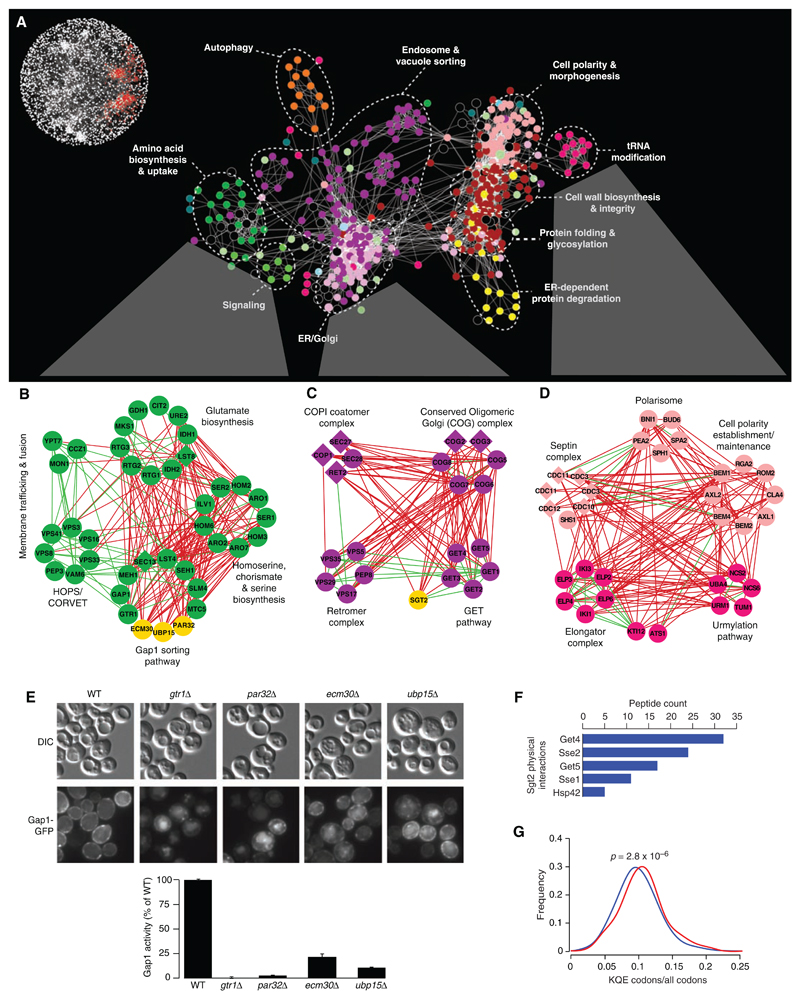
Fig. 2.
Magnification of the functional map better resolves cellular processes. (A) A subnetwork corresponding to a region of the global map described in Fig. 1 is indicated in red (inset). Node color corresponds to a specific biological process: dark green, amino acid biosynthesis and uptake; light green, signaling; light purple, ER-Golgi; dark purple, endosome and vacuole sorting; yellow, ER-dependent protein degradation; red, protein folding and glycosylation, cell wall biosynthesis and integrity; fuchsia, tRNA modification; pink, cell polarity and morphogenesis; orange, autophagy; and black, uncharacterized. Individual genetic interactions contributing to genetic profiles revealed by (A) are illustrated for three specific subnetworks in (B) to (D). (B to D) Subsets of genes belonging to amino acid biosynthesis and uptake, ER-Golgi, and tRNA modification regions of the network were selected, and, in some cases, additional genes were included from the complete network shown in Fig. 1. Nodes are grouped according to profile similarity, and edges represent negative (red) and positive (green) genetic interactions (|ε| > 0.08, P < 0.05). Nonessential (circles) and essential (diamonds) genes are colored according to the biological process indicated in (A), and uncharacterized genes are depicted in yellow. (E) PAR32, ECM30, and UBP15 are required for plasma membrane localization (micrographs) and activity (histogram) of the Gap1 amino acid permease. DIC, differential interference contrast; GFP, green fluorescent protein. (F) Sgt2 physically interacts with components of the GET pathway and members of the Hsp70 chaperone family. Proteins identified with high confidence as specific interactors for tandem affinity purification (TAP)–tagged Sgt2 (Sgt2-TAP) are shown in decreasing order of spectral counts. (G) Distribution of the Elp and Urm modified codon usage among synthetic sick or lethal interaction partners. The fraction of Elp and Urm modified codons (lysine, glutamine, and glutamic acid) relative to all codons was measured for all negative interactors with genes in the Elp or Urm complex (red) relative to the background usage of all genes (blue).