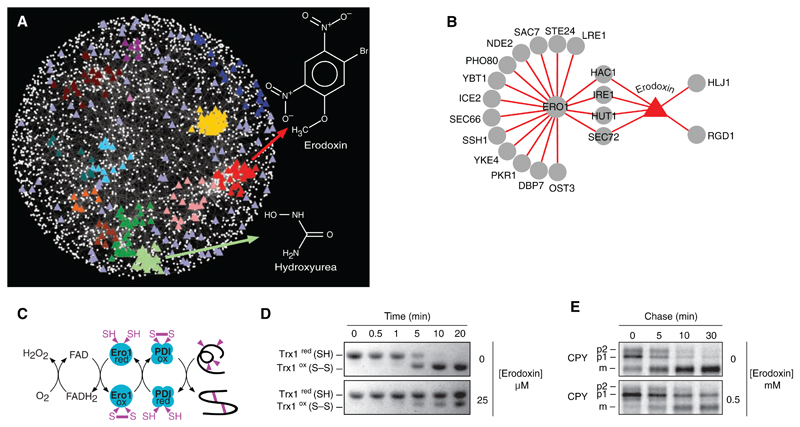
Fig. 5.
(A) A chemical-genetic interaction map is shown in which colored triangles represent chemical compounds and white nodes correspond to genes. Compounds were positioned on the map by highlighting the gene node whose genetic interaction profile most closely resembles the chemical genetic profile of the compound derived from three sources (7, 8). Compounds tightly correlated to genes positioned within functional clusters (Fig. 1) were colored accordingly to the color of the cluster as in Fig. 1. The chemical-genetic profile of hydroxyurea clustered with genes involved in DNA replication and repair, whereas that of erodoxin clustered with genes involved in protein folding, glycosylation, and cell wall biosynthesis. Compounds positioned outside functional clusters are colored light purple. (B) Network displaying overlap between ERO1 negative genetic interactions and genes resulting in growth inhibition when deleted in the presence of erodoxin. (C) ERO1-dependent pathway for oxidative protein-folding pathway. (D) Erodoxin inhibits Ero1-dependent oxidation of Trx1 in vitro. (E) Erodoxin inhibits CPY processing to the vacuolar form in vivo. ER (p1), Golgi (p2), and vacuolar (m) forms of CPY are indicated.