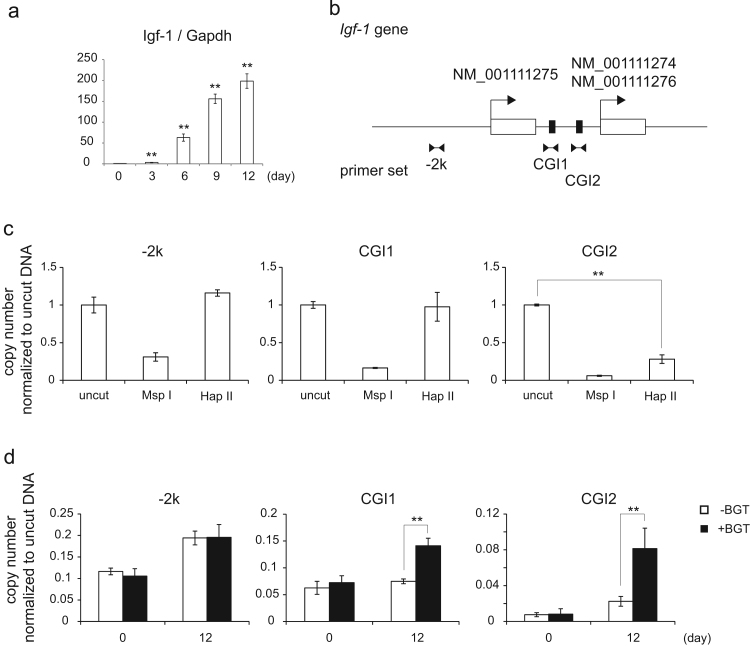
Fig. 4.
The hydroxylation of methylated DNA of the Igf1 promoter in chondrocyte differentiation. (a) RT-qPCR was carried out using a primer for Igf1 as in Fig. 1(f). Data present means ± standard deriviations (n=3). ** p<0.01. (b) Schematic representation of the Igf1 promoter. There are two transcription start sites (arrows) for NM_001111275, and NM_001111274/6. The Igf1 promoter has two CpG islands (solid boxes) between the first exons (empty boxes). For the following experiments, primers were designed for 2,000 base pairs upstream from the transcription start site (−2k) and at two CpG islands (CGI1 and CGI2). (c) Genomic DNA derived from undifferentiated C3H10T1/2 cells was digested by MspI or HapII or not treated. qPCR was conducted using −2k, CGI1 and CGI2 primers. qPCR intensity was normalized to uncut DNA samples. Data present means ± standard deriviations (n=3). **p<0.01 (d) DNA was extracted from undifferentiated cells (day 0) or differentiated cells (day 12) and was reacted with/without BGT. The glycosylated DNA was digested by MspI and qPCR was conducted as in (c). Data present means ± standard deriviations (n=3). **p<0.01.