Figure 1. Rct1 is Required for siRNA Biogenesis and Pericentromeric Heterochromatin Silencing, See also Table S1, Figures S1 and S2.
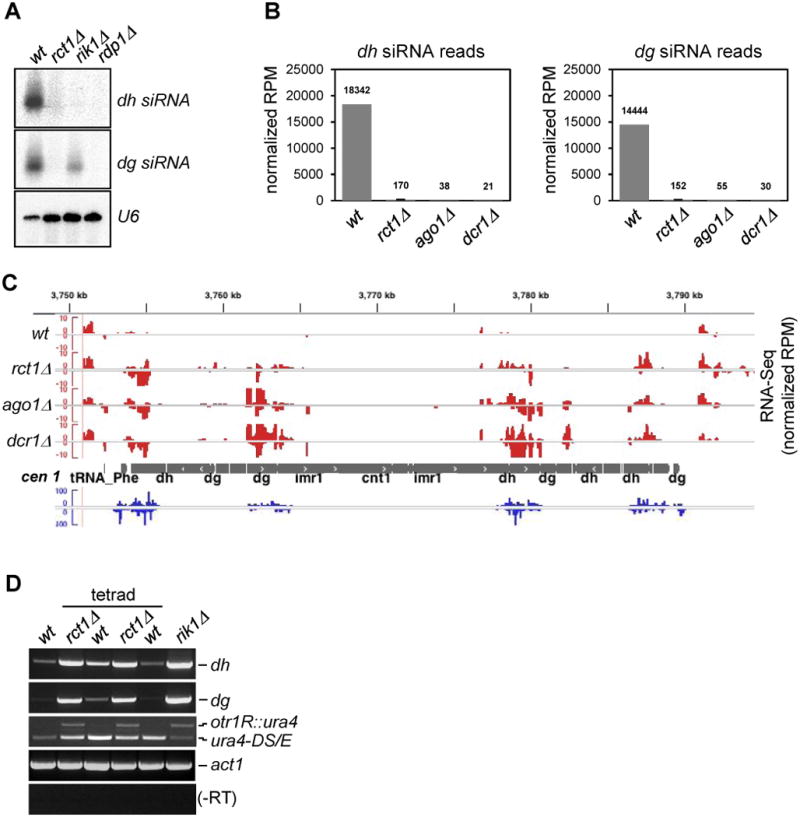
(A) Small RNA northern blots of pericentromeric dh/dg derived siRNAs. U6 serves as loading control.
(B) Quantification of cen siRNAs in indicated strains. Y-axis represents normalized reads in each library (read per million). Normalized reads mapped to dh/dg repeats are plotted separately. Data from two biological replicates of rct1Δ mutant cells were analyzed. Error bar indicates standard error from mean (SEM).
(C) RNA-seq reads at centromere (cen) 1 from indicated strains. RNA-seq tracks (red), small RNA-seq track (blue), cen 1 (grey). Y-axis represents normalized reads in each library (RPM). Only selected features are annotated on Integrated Genome Viewer screenshots in this and all subsequent figures.
(D) Semi-quantitative RT-PCR of dh/dg and otr1R::ura4 transcript levels in rct1Δ mutant cells. Truncated ura4-DS/E at the endogenous site and act1 serves as loading controls, -RT omits the reverse transcription step. A full tetrad was analyzed to show that the silencing defect phenotype segregates with rct1Δ alleles.
