Abstract
Cell-cell signaling between bacteria, including quorum-sensing (QS) communication systems, may play a role in the establishment and maintenance of polymicrobial communities. To better understand and model these interactions, we must uncover the degree to which neighboring species recognize each another’s signals. In the current study, we tested the likelihood of whether the QS systems of two opportunistic pathogens (Acinetobacter baumannii and Pseudomonas aeruginosa) that frequently arise in polymicrobial infections would be affected by the QS signals of neighboring species. Through the synthesis and screening of a library of native and non-native N-acyl l-homoserine lactones (AHLs), we found that the AbaR LuxR-type receptor protein of A. baumannii is highly selective for its native AHL signal. However, a homologous LuxR-type receptor in P. aeruginosa, LasR, is far more promiscuously activated by AHLs relative to AbaR, suggesting that LasR-regulated QS could be more susceptible to activation by neighboring species. To explain the observed difference in signal selectivity between AbaR and LasR, we developed a model based on (i) the activity profiles of these proteins and (ii) previously reported structural data and activity profiles for related LuxR-type receptors. This model may facilitate the study of signal selectivities for hundreds of LuxR-type QS receptors from bacteria, many of which grow in polymicrobial communities and may sense each other’s signals. In addition, we discovered a set of AHLs that could be used to selectively activate LasR and selectively inhibit AbaR in polymicrobial experiments.
Graphical Abstract
INTRODUCTION
Most microbes grow in communities containing multiple species that cooperate and compete for optimal fitness.1 Such polymicrobial communities can have a widespread influence on human life and livelihood. For example, bacterial infections are often caused by a mixture of pathogenic species,2 and polymicrobial biofilms can play beneficial and detrimental roles in industry and the environment.3–6 We are also increasingly appreciating the importance of the human microbiome for both positive and negative health outcomes.2, 7
In these mixed microbial communities, microbes secrete different factors (e.g., small molecules, peptides, and proteins) into the environment that have the potential to affect other community members. These interactions between microbes can result in significant benefit or harm to the local environment or to host organisms.8 Computational modeling methods show promise at describing and predicting polymicrobial interactions; however, these models require more empirical data reporting the molecular details of microbial interactions.9 One manner in which microbes can interact is via quorum sensing (QS).10, 11 Bacterial QS is generally viewed as a mechanism by which bacteria sense their population density and alter their behavior for optimal fitness under that density (although other explanations for the phenomenon exist12). In QS, bacteria synthesize a small molecule or peptide signal that can diffuse or is secreted into the environment. A threshold density of bacteria is required for the signal to accumulate at a concentration sufficient for productive binding of cognate receptor proteins in the bacterial cells. This binding event subsequently induces changes in gene expression necessary for the initiation of a range of behaviors that benefit the bacterial population at high density. In the case of the Gram-negative Proteobacteria phylum, N-acyl l-homoserine lactone (AHL) QS signals are biosynthesized by LuxI-type enzymes and sensed by receptor proteins of the LuxR-type transcriptional regulator class.10 Significantly, the group behaviors regulated by AHL-based QS are often tied to virulence in pathogens and to biofilms in a wide range of species.11
AHL-based QS is generally considered a method for intraspecies bacterial communication.10, 11 However, since many Gram-negative bacteria that utilize QS live in close proximity (e.g., on eukaryotic hosts and in the soil), it is likely that their AHL signals could activate or disrupt the QS systems of other bacteria and potentially “coerce” them to initiate different behaviors,13 or conversely, bacteria could “eavesdrop” on their neighbors and modify their own behavior appropriately.14 For example, the opportunistic pathogen Pseudomonas aeruginosa can activate the QS system of Burkholderia cepacia in mixed biofilms in cystic fibrosis lung infection models.15 As such, examining the mechanisms and scope of bacteria using QS for interspecies—and even interkingdom—sensing is an area of significant and increasing research interest.16, 17
Current genomic technologies allows for the rapid identification of microorganisms present in a given environment and the likely communication genes that they possess.18 However, tools to discover the actual degree of chemical cross-talk between species within a population and the mode by which that cross-talk influences the population’s behavior are limiting. The development of species-selective, small molecule QS modulators to regulate the QS of individual species within a complex polymicrobial mixture would allow for the interrogation of the impact of QS cross-talk on the behavior of the communities in native environments. The development of such chemical tools is a long-term goal of ours and a broad motivation for the current study.
Herein, we report our investigations into the selective modulation of two LuxR-type receptor proteins from two different bacteria, AbaR of Acinetobacter baumannii and LasR of P. aeruginosa, using AHL-type ligands. There were three main objectives for this research. First, we aimed to comprehensively characterize the selectivities of AbaR and LasR for their native ligand relative to other naturally occurring AHLs. Second, we sought to discover AHL-type ligands that can serve as chemical probes to selectively activate or inhibit the QS systems of AbaR or LasR; such tools could allow for the orthogonal modulation of their QS systems in polymicrobial communities. Third, we aimed to use our uncovered ligand-promiscuity data, along with prior small molecule screening data in other LuxR homologs, to develop a mechanistic model to explain the promiscuity differences between AbaR and LasR on a molecular level. We realized each of these three research objectives, obtaining results that should shape future studies of the selectivity and cross-talk between LuxI/LuxR-type QS systems in other co-localized bacteria
RESULTS AND DISCUSSION
Rationale for receptor selection
Two main reasons prompted the choice of AbaR and LasR for focused study. First, an initial study by our laboratory suggested that these two receptors have different degrees of selectivity for their native signal molecules,19 even though their native AHL signals are very similar: LasR naturally responds to N-(3-oxo-dodecanoyl) l-homoserine lactone (OdDHL, ligand 1 in Fig. 1) to regulate numerous virulence phenotypes in P. aeruginosa,20 and AbaR naturally responds to (R)-N-(3-hydroxydodecanoyl) l-homoserine lactone ((R)-OH-dDHL, ligand 2 in Fig. 1) to regulate surface motility and biofilm formation (putatively linked to virulence) in A. baumannii.21, 22 Therefore, we reasoned that studying these proteins could yield explanations to which features makes a LuxR-type protein more selective for a given AHL signal over others. Second, P. aeruginosa and A. baumannii reside in polymicrobial contexts where interspecies cross-talk is possible. These two species are widespread in the environment (e.g., in soil and water), where they live in proximity to multitudes of other microbes, and they are each opportunistic pathogens of immunocompromised humans, where they often form polymicrobial infections7, 23 (and sometimes with each other24).
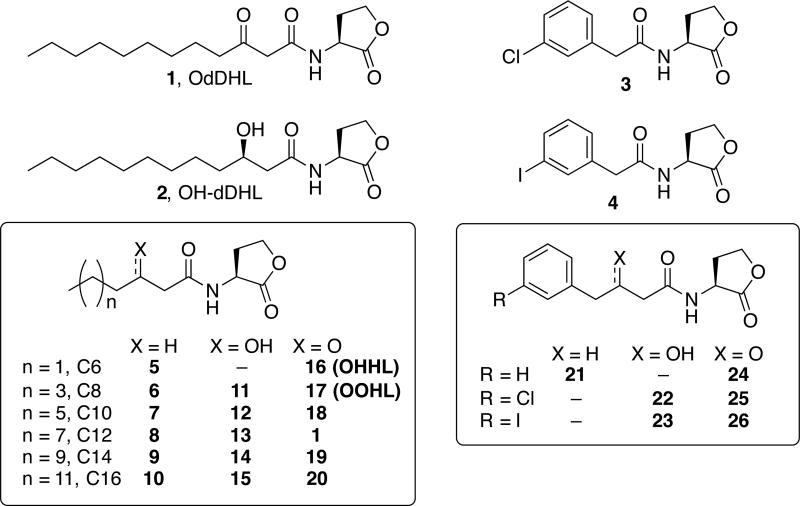
Fig. 1.
The natural AHL ligands for LasR (1, OdDHL) and AbaR (2, (R)-OH-dDHL), and the other AHLs evaluated in this study (3–26).
Library design and synthesis
To characterize the ligand selectivities of AbaR and LasR and to identify chemical tools for their differential activation, we assembled and screened a focused library of 24 AHL-type ligands (3–26) for activation and inhibition of AbaR and LasR (Fig. 1). As highlighted above, the native AHLs of LasR and AbaR (1 and 2, respectively) are very similar, only differing in oxidation state at the 3-posititon on their acyl chain. To systematically test the impact of acyl chain length and 3-position oxidation state on activity in AbaR and LasR, we examined aliphatic AHL ligands 5–20 (Fig. 1), of which 9, 11–15, and 20 represent new additions to our growing AHL libraries.25 (We have performed initial studies on the remaining AHL ligands in LasR and AbaR (see Table S1), but we reevaluated these compounds here under the same experimental conditions as the new ligands to facilitate legitimate data comparisons).19, 26, 27 AHLs 5–20 were synthesized using standard methods (see SI); the hydroxyl-containing ligands 11–15 were prepared as diastereomeric mixtures, whereas ligand 2 is the (R)-OH-dDHL diastereomer. Many of these aliphatic-tail AHLs are, in fact, naturally occurring signal molecules for various soil-dwelling Proteobacteria (e.g., 5, N-hexanoyl l-homoserine lactone in Chromobacterium violaceum; 16, N-(3-oxo-hexanoyl) l-homoserine lactone (OHHL) in Pectobacterium carotovora; and 17, N-(3-oxo-ocatanoyl) l-homoserine lactone (OOHL) in Agrobacterium tumefaciens; see Table S2 matching other bacteria to their cognate signals). Therefore, we reasoned that testing the abilities of these AHLs to modulate the AbaR and LasR QS receptors would likely not only provide new chemical probes, but also could illuminate the likelihood of A. baumannii and P. aeruginosa to be affected by cross-talk with neighboring Proteobacteria in real polymicrobial communities.
Since many of our previously reported, highly active non-native AHLs contain substituted phenyl groups in the acyl tail (e.g., ligands 3 and 4, Fig. 126, 27) and again, LasR and AbaR respond to native AHL ligands that only differ in oxidation state at the 3-position (1 and 2), we sought to examine the effect of altering the 3-position oxidation state of phenyl-substituted AHLs on their activity profiles in AbaR and LasR. Therefore, we examined the N-(phenylbutanoyl) l-homoserine lactone (PBHL) ligands 21–26, which have unsubstituted or chlorine- or iodine-substituted phenyl groups and varying 3-position oxidation states (Fig. 1). Within this set of compounds, PBHLs 22, 23, and 26 constitute new members of our in-house AHL libraries (see Table S1) and were synthesized using established procedures (22 and 23 were diastereomeric mixtures; see SI).
AbaR is highly selective for aliphatic-tail AHLs with 12-carbon chains and 3-position hydroxyls, while LasR is modulated more promiscuously by AHLs
As our initial studies suggested that AbaR was particularly sensitive to AHL acyl chain length and oxidation state,19 we sought to more rigorously characterize the limits whereby AHL acyl chain lengths and oxidation states would activate or inhibit AbaR activity, and thereafter, directly compare these limits to those for LasR activation and inhibition. We thus began our studies by examining the abilities of the aliphatic AHLs 5–20 to either activate or inhibit AbaR and LasR using bacterial reporter strains (see SI for full assay details). In these assays, activated AbaR and LasR induce the production of the enzyme β-galactosidase, the amount of which can be quantified by its cleavage of a colorimetric substrate. Activation (or agonism) was tested by adding an AHL, while competitive inhibition (or antagonism) was tested by adding native ligand near its EC50 (concentration that elicits a half-maximum activation) along with a non-native AHL of interest. The primary assay data resulting from these screens are shown in Fig. 2 and Table S3, the latter of which highlights the dependence of AbaR and LasR activity on acyl chain length and oxidation state. For ligands displaying sufficient activity, we also analyzed their potencies by determining their EC50 or IC50 (concentration that elicits a half-maximum inhibition) in the same reporter assays.
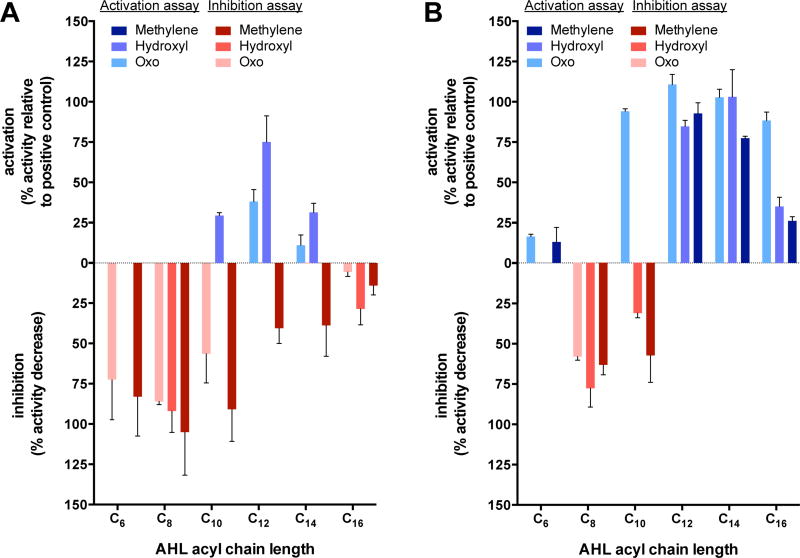
Fig. 2.
Comparative activation and inhibition data for AbaR (A) and LasR (B) by aliphatic AHLs. Activation is reported in blue shades on the positive y-axis as the % activity relative to that induced by the receptor’s native ligand (2 for AbaR and 1 for LasR). Inhibition activity is reported in red shades on the negative y-axis as the % decrease in activity relative to only native ligand being present. When both activation and inhibitory activity were observed, only the activity with the highest percent value is displayed for clarity. Activities of 3-methylene AHLs 5–10 displayed with dark shades, activities of 3-hydroxyl AHLs 11–15 displayed with medium shades, and activities of fully oxidized 3-oxo AHLs 16–18, 1, 19, and 20 displayed with light shades. Error bars represent s.e.m. of a biological triplicate. See Table S3 legend and Supplemental Methods for assay details.
The agonism assay data indicated that AbaR was far more selectively activated by aliphatic AHLs (5–20) relative to LasR (Fig. 2), confirming our initial predictions. All lengths of the fully reduced, or “3-methylene”, AHLs (5–10) were incapable of activating AbaR, and most were instead strong competitive inhibitors of AbaR activation by its native ligand (2) (see Fig. 2A, dark shades). In particular, 3-methylene AHLs with chain lengths below 12 carbons (5–7) were the strongest inhibitors. “Fully oxidized”, or 3-oxo, ligands were slightly better at activating AbaR than the 3-methylene AHLs; however, only the 12-carbon 3-oxo AHL, OdDHL (1), was capable of appreciable AbaR agonism (~50% relative to activation by the same concentration of AbaR’s native ligand, (R)-OH-dDHL (2)). All other chain lengths of the 3-oxo AHLs were incapable of significant AbaR activation, and in fact, the shorter chain (i.e., C6, C8, and C10) ligands 16–18 were good competitive inhibitors of AbaR instead (Fig. 2A, light shades). The hydroxyl, or 3-OH, AHL class was the best at activating AbaR, which is not surprising given that its native ligand (2) bears a 3-OH moiety (Fig. 2A, medium shades).
The mixture of diastereomers present in ligand 13 demonstrated weaker activity than the diastereomerically pure native ligand (R)-OH-dDHL (2). This result suggests that the non-native diastereomer is a partial agonist of AbaR. Although other possibilities exist, we suspect that the non-native diastereomers present in ligands 11, 12, 14, and 15 are also partial agonists, which would account for a moderate reduction in the activation by these ligands. Nonetheless, in these diastereomeric mixtures, only the 3-OH C10 and C14 ligands activated AbaR by more than 25%—a low threshold for activity. At longer and shorter chain lengths, the hydroxyl AHLs failed to activate AbaR, but instead were competitive inhibitors—especially the shorter C8 ligand 11. In total, these screening data suggest that AbaR can only be activated by aliphatic AHLs that vary slightly from its native ligand (2) in either oxidation state or chain length, but not both. Most of the aliphatic AHLs tested, especially those with shorter chains, were instead good competitive inhibitors of AbaR.
In contrast, the LasR reporter assays revealed that LasR was much more promiscuously activated than AbaR by AHLs 5–20 (Fig. 2, Table S3). AHLs with any of the three oxidation states (3-methylene, 3-OH, or 3-oxo) at the native-like C12 length (8, 13, and OdDHL (1)) were able to substantially activate LasR (>75% relative to activation by the same concentration of LasR’s native ligand (1)). The 3-methylene and 3-OH ligands were still good agonists (>75%) at the C14 length (9, 14), but were much weaker agonists at the C16 length (10, 15) and at chain lengths less than C12. The 3-methylene and 3-OH ligands of both lengths C8 (6, 11) and C10 (7, 12) were competitive inhibitors of LasR, instead. LasR was activated by an even wider range of ligands with the native 3-oxo functionality: chain lengths of C10 through C16 (1, 18, 19, and 20) all activated LasR by >75%. Only the C8 3-oxo ligand 17 was a competitive LasR inhibitor. Together, these reporter assay results suggest that LasR can be activated by aliphatic AHLs of substantial variation in chain length or oxidation state, and can even tolerate ligands with variations in both traits.
In view of these data for AHLs 5–20, it is obvious that both AHL acyl chain oxidation state and length play key roles for modulation of both AbaR and LasR, but the tolerance thresholds differ substantially. With regard to the biological relevance of these activity profiles, it is quite interesting that AbaR is far less promiscuously activated than LasR. As highlighted earlier, both A. baumannii and P. aeruginosa are increasingly found in mixed microbial environments, where other species can be producing AHLs with various lengths and oxidation states, the bulk of which are included in the aliphatic AHL sub-library tested here (5–20).7, 23, 24 However, our data suggest that these two bacteria may have opposite approaches to respond to the AHLs of other species: A. baumannii QS is not activated—but in fact most commonly inhibited—by these foreign signals, whereas P. aeruginosa QS is activated. The fitness implications of these two strategies are an exciting avenue for future research,13 and we return to a potential biochemical rationale for this relative selectivity and promiscuity below.
AbaR and LasR are inhibited to similar degrees by aromatic AHLs with different 3-position oxidation states
We next examined the effects of altering the 3-position oxidation state in the non-native PBHL ligands (21–26) on ligand activity in AbaR and LasR using the analogous reporter strain assays as described above. As shown in Table 1, all six PBHLs are inhibitors of AbaR, and all but ligand 21 are inhibitors of LasR. None of the PBHLs demonstrated appreciable agonist activity on either receptor (see Fig. S1 and Fig. S6). Similarly, the N-(phenylacetanoyl) l-homoserine lactones 3 and 4, which are two-carbon-shorter analogs of 22/25 and 23/26, respectively, were found to be inhibitors of both LasR (40–60%)27 and AbaR (70–90%)19 (see also Fig. S2 and Fig. S4). The AbaR data indicate that the 3-position oxidation state has no impact on the ability of these PBHLs to modulate AbaR (i.e., 3-methylene 21 was similar to 3-oxo 24, 3-OH 22 was similar to 3-oxo 25, and 3-OH 23 was similar to 3-oxo 26). For LasR however, the ligand oxidation state impacted inhibition somewhat, as the percent inhibition value for 3-oxo 24 was higher than 3-methylene 21, and 3-OH 22 inhibited LasR more than 3-oxo 25. Additionally, the IC50 value for 3-oxo 26 was > 4× lower than that for 3-OH 23 with LasR. However, these effects were moderate, and there was no obvious trend to suggest that a specific oxidation state yielded the strongest inhibitory activity for this ligand class against LasR. In sum, AbaR inhibition did not depend significantly on the oxidation state of the PBHL ligand, and LasR inhibition was only moderately dependent on PBHL oxidation states.
Table 1.
AbaR and LasR primary antagonism assay data and IC50 values with confidence intervals for the aromatic-tail AHLsa, b
| AbaR | LasR | |||
|---|---|---|---|---|
| AHL | Inhibition (%)c | IC50 value (nM)d | Inhibition (%)c | IC50 value (µM)d |
| 21 | 56 | 0 | ||
| 22 | 74 | 7.54 (6.81–8.34) | 89 | 1.50 (0.778–2.89) |
| 23 | 73 | 7.36 (5.74–9.44) | 48 | 1.98 (1.26–3.10) |
| 24 | 51 | 20 | 1.91 (0.885–4.10) | |
| 25 | 51 | 10.5 (7.16–15.4) | 53 | 1.53 (0.734–3.17) |
| 26 | 69 | 20.0 (9.28–43.0) | 54 | 0.458 (0.191–1.10) |
All assays performed in biological triplicate.
All ligands were screened for agonism of LasR and AbaR, yet none showed appreciable agonist activity. See SI.
AHLs evaluated at 100 µM against 2 at 700 nM for AbaR or at 10 µM against 1 at 10 nM for LasR. Error ≤ ±10%.
IC50 values determined by testing AHLs over a range of concentrations against 2 at 700 nM for AbaR or 1 at 10 nM for LasR.
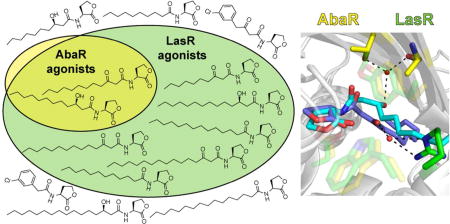
Collectively, screening of this library of alkyl and aryl AHLs, despite its small size, has provided a set of ligands with a range of different activity and selectivity profiles in AbaR and LasR. These compounds can activate both AbaR and LasR (13), inhibit both AbaR and LasR (6, 7, 11, 17, 22, 23, 25, 26), activate LasR only (19, 20), inhibit AbaR only (5, 16, 21), or both activate LasR and inhibit AbaR at the same time (18). This study only failed to reveal ligands that selectively activate AbaR and/or selectively inhibit LasR. Nevertheless, other than some of our past studies,26–29 the development of ligands with LuxR-type receptor selectivity remains largely unchartered; thus, the discovery of these activity profiles for these ligands in AbaR and LasR is significant. These ligands could be utilized in a range of interesting experiments of both fundamental and direct clinical interest to test the impact of AbaR and LasR modulation in mixed cultures of A. baumannii and P. aeruginosa—mixtures that are relevant to infection populations.24
A model for the different selectivities of LasR and AbaR for their native AHL signals
From the reporter assay data above, it appears that the primary difference in AbaR:AHL and LasR:AHL structure activity relationships (SARs) is that AbaR was only activated by >50% by its native ligand, whilst ligands of multiple oxidation states and chain lengths were good LasR activators. We aimed to develop a plausible model for the ligand selectivity of AbaR.
We began by scrutinizing X-ray crystal structures of LuxR-type proteins to garner insights into ligand-binding selectivities. Although no solid-state structure of AbaR has been reported, structures have been solved for the ligand-binding domain of LasR and two other full length LuxR-type proteins that natively bind AHLs with polar moieties at the 3-position (i.e., TraR from A. tumefaciens and the orphan receptor QscR from P. aeruginosa).30–32 Notably, these three receptors present three different modes of binding the 3-oxo moiety of their native AHL ligand (or preferred ligand for QscR) (Fig. 3A,B,C). We and others have compiled substantial SARs for both native and non-native ligands with TraR, LasR, and QscR,19, 27, 33–35 and a correlation is observed between apparent receptor selectivity for activation by AHL-type ligands (as determined in reporter assays) and the mode in which these receptors bind the AHL 3-oxo group (as determined by structural analyses). For instance, TraR is far more discriminately activated than LasR.27, 36 This selectivity can be explained, at least in part, by the observation that the 3-oxo moiety of TraR’s native ligand, OHHL, forms a hydrogen bond simultaneously with two TraR residues (the Thr129 side-chain and Asp38) (Fig. 3A).32 Therefore, a ligand change would likely require a greater restructuring of the TraR protein compared to LasR, in which only one side chain (Arg61)30 would be immediately affected (Fig. 3A, B). In contrast, prior studies have revealed that QscR is more promiscuously activated than LasR.28, 33 Churchill and coworkers noted that in their X-ray crystal structure of QscR with OdDHL (1), the ligand’s 3-oxo moiety hydrogen bonds with the QscR’s Ser56 through a chain of two water molecules, thereby imparting greater flexibility of ligand binding (Fig. 3C).31 (We note that X-ray crystal structures of SdiA [an orphan LuxR-type receptor from E. coli] bound to 3-oxo ligands OHHL (16) and OOHL (17) have been reported recently as well; however, conflicting activity data37–39 and multiple orientations of the ligands in the structures40 preclude a comparative analysis of SdiA with AbaR and LasR here.)
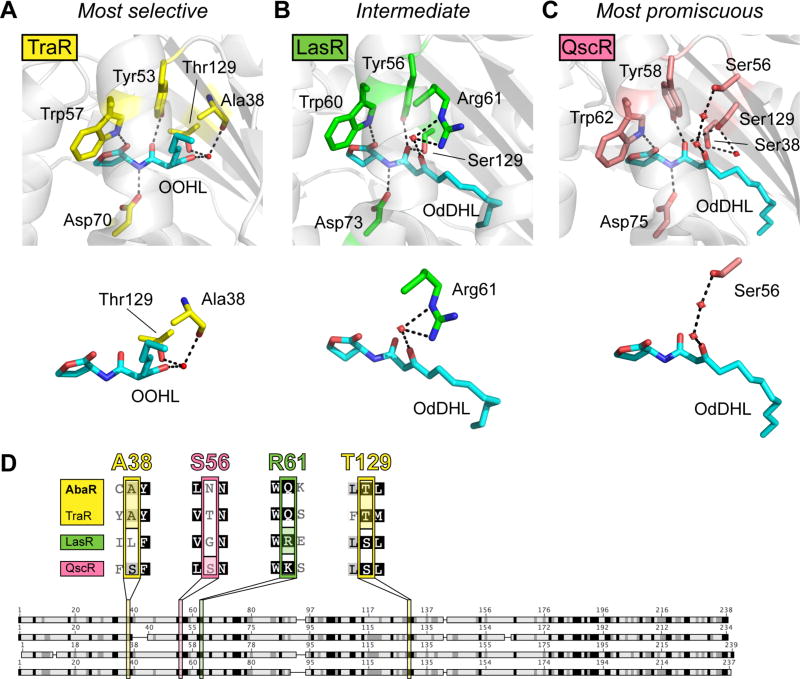
Fig. 3.
Model to explain AHL-selectivity in AbaR and LasR. (A) Ligand-binding pocket of X-ray crystal structure of TraR-OOHL (pdb 1L3L).32 (B) Ligand-binding pocket of X-ray crystal structure of LasR-OdDHL (pdb 2UV0).30 (C) Ligand-binding pocket of X-ray crystal structure of QscR-OdDHL (pdb 3SZT).31 Residues that form hydrogen bonds with the ligand are displayed on top, and simplified versions of binding pocket images displaying only the residues involved in hydrogen-bonding with the 3-oxo ligand moiety are shown below. Black dashed lines indicate hydrogen bonds, and red spheres indicate water molecules. (D) Alignment of AbaR with three 3-oxo-AHL-binding LuxR homologs. See Table S4 for full alignment. Darker shading indicates higher degree of conservation. Four residues are highlighted that bind the 3-oxo moiety of the proteins’ native AHL in reported X-ray crystal structures (A38 and T129 for TraR, S56 for QscR, and R61 for LasR).
We lack structural data for AbaR for direct comparison to LasR, TraR and QscR. However, we reasoned that analysis of protein sequence data could provide insights into which receptor binds its ligand most like AbaR. We therefore performed a sequence alignment analysis for LasR, TraR, QscR, and AbaR (Fig. 3D and Table S4). When the amino acid sequence of AbaR’s ligand-binding region is compared to that of the other receptors (Fig. 3D), it is apparent that AbaR shares the TraR 3-oxo-binding residues Thr129/Ala38—not the LasR or QscR 3-oxo-binding residues (Arg61 in LasR is Glu in AbaR; Ser56 in QscR is Asn in AbaR). This observation is congruent with our activity data indicating that both AbaR and TraR are selective for their native ligands. We therefore propose that AbaR may bind its native ligand (R)-OH-dDHL (2) in a similar mode to TraR for OOHL (17), and that this binding mode is more selective for a specific cognate AHL than is the ligand binding pocket of LasR or QscR. The ensemble of additional interactions between the receptor proteins and their ligands (e.g., interactions with the ligand’s hydrophobic tail31, 36) are certain to further refine the differential selectivities of these three proteins. Future structural studies of the AbaR:OH-dDHL complex would offer critical insight into these additional key contacts, as well as test the hypothesized 3-hydroxyl moiety binding by Thr129/Ala38. More broadly, analogous SAR and alignment studies of LuxR homologs in other bacteria would supplement this finding by testing the generality of our observed ligand promiscuity-sequence correlation reported here for LasR, TraR, QscR, and AbaR.
Summary and outlook
This study was motivated by our broad interest in interspecies, AHL-based signaling in mixed microbial environments. Herein, we report a systematic characterization of the impact of AHLs with different acyl chain lengths, 3-position oxidation states, and gross structures (aliphatic versus aromatic) on activation and inhibition of the AbaR and LasR receptor proteins from A. baumannii and P. aeruginosa. We found that AbaR in A. baumannii is selective for its native ligand, whilst LasR in P. aeruginosa is far more tolerant of AHL structural changes. Out of this focused library of AHLs, AbaR was only activated >50% by its native ligand (R)-OH-dDHL (2), and only minimal changes (i.e., 3-OH to 3-oxo conversion, or two-carbon additions or reductions to its acyl chain) could be tolerated to maintain any activation at all. These data demonstrate that A. baumannii QS should not be activated by the AHL signals of many neighboring bacteria (in contrast to P. aeruginosa, which has more promiscuous LuxR-type receptors; e.g., LasR and QscR). It is intriguing to speculate that over the course of evolutionary history, AbaR may have undergone a selection pressure that gave its host bacterium an advantage by ignoring (or at least not being activated by) the AHLs produced by neighboring bacteria. In contrast, LasR possibly had no such selective pressure—or even conferred a selective advantage to its host bacterium by responding to a broader AHL range.
This study also revealed a set of AHL ligands that could be useful chemical probes to activate both AbaR and LasR, inhibit both of these receptors, selectively activate LasR, selectively inhibit AbaR, or both activate LasR and inhibit AbaR. These compounds display a remarkable level of diversity in their activity profiles, and represent valuable new probes to study QS in A. baumannii and P. aeruginosa alone, or perhaps most interestingly, in mixed cultures. Lastly, by analyzing sequence alignments and X-ray crystal structures, we proposed a molecular rationale for the higher selectivity of AbaR compared to LasR. The deeper understanding of the A. baumannii and P. aeruginosa QS systems afforded by this study is significant because virulence phenotypes of both of these pathogens are regulated by QS,19–22 and both pathogens are of significant clinical concern due to their propensity to antibiotic resistance, being prominent members of the ESKAPE pathogen group.41 Moreover, and aligned with the broad motivation for this study, these bacteria have both been observed extensively in polymicrobial infections where QS cross-talk could likely occur. Looking to the future, we expect the analysis of LuxR homolog promiscuity presented in this work and proposed previously by others31, 32 could have broader applications beyond A. baumannii and P. aeruginosa. As the molecular rationales for receptor selectivity become more refined, they may eventually lead to sequence-based predictions of QS receptor promiscuity in other polymicrobial communities.
Supplementary Material
Acknowledgments
The NIH (GM109403) and Burroughs Wellcome Fund provided generous financial support for this work. D.M.S. was funded in part by an NSF Graduate Fellowship (DGE-0718123). J.P.G. was funded in part by a National Defense Science & Engineering Graduate Fellowship (32 CFP 168a) and by the NIH through the UW–Madison Chemistry-Biology Interface Training Grant (T32 GM008505). M.E.B. was funded in part by the NSF through the UW–Madison Materials Research Science and Engineering Center (DMR-1121288). NMR facilities in the UW–Madison Department of Chemistry were supported by the NSF (CHE-0342998) and a gift from P. Bender. MS facilities in the UW–Madison Department of Chemistry were supported by the NSF (CHE-9974839). We thank P. Rather (Emory University) for the A. baumannii strain used in this study.
Footnotes
Electronic supplementary information (ESI) available
Full experimental methods, table of AHLs used by selected bacteria, primary screening data, dose response curves, full sequence alignment, and characterization data for all new compounds.
References
- 1.Hibbing ME, Fuqua C, Parsek MR, Peterson SB. Bacterial competition: surviving and thriving in the microbial jungle. Nat. Rev. Microbiol. 2009;8:15–25. doi: 10.1038/nrmicro2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burmølle M, Ren D, Bjarnsholt T, Sørensen SJ. Interactions in multispecies biofilms: do they actually matter? Trends Microbiol. 2014;22:84–91. doi: 10.1016/j.tim.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 3.Lenhart TR, Duncan KE, Beech IB, Sunner JA, Smith W, Bonifay V, Biri B, Suflita JM. Identification and characterization of microbial biofilm communities associated with corroded oil pipeline surfaces. Biofouling. 2014;30:823–835. doi: 10.1080/08927014.2014.931379. [DOI] [PubMed] [Google Scholar]
- 4.Meyer A, Wallis FM. Development of microbial biofilms on various surfaces for the treatment of heavy metal containing effluents. Biotech. Tech 1997 [Google Scholar]
- 5.Stoodley P, Sauer K, Davies DG, Costerton JW. Biofilms as complex differentiated communities. Annu. Rev. Microbiol. 2002;56:187–209. doi: 10.1146/annurev.micro.56.012302.160705. [DOI] [PubMed] [Google Scholar]
- 6.Zimmer BL, May AL, Bhedi CD, Dearth SP, Prevatte CW, Pratte Z, Campagna SR, Richardson LL. Quorum sensing signal production and microbial interactions in a polymicrobial disease of corals and the coral surface mucopolysaccharide layer. PLoS ONE. 2014;9:e108541–108516. doi: 10.1371/journal.pone.0108541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peters BM, Jabra-Rizk MA, O'May GA, Costerton JW, Shirtliff ME. Polymicrobial interactions: impact on pathogenesis and human disease. Clin. Microbiol. Rev. 2012;25:193–213. doi: 10.1128/CMR.00013-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shank E, Kolter R. New developments in microbial interspecies signaling. Curr. Opin. Microbiol. 2009 doi: 10.1016/j.mib.2009.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Widder S, Allen RJ, Pfeiffer T, Curtis TP, Wiuf C, Sloan WT, Cordero OX, Brown SP, Momeni B, Shou W, Kettle H, Flint HJ, Haas AF, Laroche B, Kreft J-U, Rainey PB, Freilich S, Schuster S, Milferstedt K, van der Meer JR, Groβkopf T, Huisman J, Free A, Picioreanu C, Quince C, Klapper I, Labarthe S, Smets BF, Wang H, Fellows INI, Soyer OS. Challenges in microbial ecology: building predictive understanding of community function and dynamics. ISME J. 2016;10:2557–2568. doi: 10.1038/ismej.2016.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fuqua C, Parsek MR, Greenberg EP. Regulation of gene expression by cell-to-cell communication: acyl-homoserine lactone quorum sensing. Annu. Rev. Genet. 2001;35:439–468. doi: 10.1146/annurev.genet.35.102401.090913. [DOI] [PubMed] [Google Scholar]
- 11.Rutherford ST, Bassler BL. Bacterial quorum sensing: its role in virulence and possibilities for its control. CSH Perspect. Med. 2012;2:a012427. doi: 10.1101/cshperspect.a012427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.West SA, Winzer K, Gardner A, Diggle SP. Quorum sensing and the confusion about diffusion. Trends Microbiol. 2012;20:586–594. doi: 10.1016/j.tim.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 13.Diggle SP, Gardner A, West SA, Griffin AS. Evolutionary theory of bacterial quorum sensing: when is a signal not a signal? Phil. Trans. R. Soc. B. 2007;362:1241–1249. doi: 10.1098/rstb.2007.2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Joint I, Allan Downie J, Williams P. Bacterial conversations: talking, listening and eavesdropping. An introduction. Phil. Trans. R. Soc. B. 2007;362:1115. doi: 10.1098/rstb.2007.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Riedel K, Hentzer M, Geisenberger O, Huber B, Steidle A, Wu H, Høiby N, Givskov M, Molin S, Eberl L. N-acylhomoserine-lactone-mediated communication between Pseudomonas aeruginosa and Burkholderia cepacia in mixed biofilms. Microbiology. 2001;147:3249–3262. doi: 10.1099/00221287-147-12-3249. [DOI] [PubMed] [Google Scholar]
- 16.Ng W-L, Bassler BL. Bacterial quorum-sensing network architectures. Annu. Rev. Genet. 2009;43:197–222. doi: 10.1146/annurev-genet-102108-134304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Teplitski M, Mathesius U, Rumbaugh KP. Perception and degradation of N-acyl homoserine lactone quorum sensing signals by mammalian and plant cells. Chem. Rev. 2011;111:100–116. doi: 10.1021/cr100045m. [DOI] [PubMed] [Google Scholar]
- 18.Kimura N. Metagenomic approaches to understanding phylogenetic diversity in quorum sensing. Virulence. 2014;5:433–442. doi: 10.4161/viru.27850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stacy DM, Welsh MA, Rather PN, Blackwell HE. Attenuation of quorum sensing in the pathogen Acinetobacter baumannii using non-native N-acyl homoserine lactones. ACS Chem. Biol. 2012;7:1719–1728. doi: 10.1021/cb300351x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schuster M, Greenberg EP. A network of networks: quorum-sensing gene regulation in Pseudomonas aeruginosa. Int. J. Med. Microbiol. 2006;296:73–81. doi: 10.1016/j.ijmm.2006.01.036. [DOI] [PubMed] [Google Scholar]
- 21.Clemmer KM, Bonomo RA, Rather PN. Genetic analysis of surface motility in Acinetobacter baumannii. Microbiology. 2011;157:2534–2544. doi: 10.1099/mic.0.049791-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Niu C, Clemmer KM, Bonomo RA, Rather PN. Isolation and characterization of an autoinducer synthase from Acinetobacter baumannii. J. Bacteriol. 2008;190:3386–3392. doi: 10.1128/JB.01929-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liao Y-T, Kuo S-C, Lee Y-T, Chen C-P, Lin S-W, Shen L-J, Fung C-P, Cho W-L, Chen T-L. Sheltering effect and indirect pathogenesis of carbapenem-resistant Acinetobacter baumannii in polymicrobial infection. Antimicrob. Agents Chemother. 2014;58:3983–3990. doi: 10.1128/AAC.02636-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dent LL, Marshall DR, Pratap S, Hulette RB. Multidrug resistant Acinetobacter baumannii: a descriptive study in a city hospital. BMC Infect. Dis. 2010;10:196. doi: 10.1186/1471-2334-10-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Welsh MA, Blackwell HE. Chemical probes of quorum sensing: from compound development to biological discovery. FEMS Microbiol. Rev. 2016;40:774–794. doi: 10.1093/femsre/fuw009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Geske GD, O'Neill JC, Miller DM, Mattmann ME, Blackwell HE. Modulation of bacterial quorum sensing with synthetic ligands: systematic evaluation of N-acylated homoserine lactones in multiple species and new insights into their mechanisms of action. J. Am. Chem. Soc. 2007;129:13613–13625. doi: 10.1021/ja074135h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Geske GD, ONeill JC, Miller DM, Wezeman RJ, Mattmann ME, Lin Q, Blackwell HE. Comparative analyses of N-acylated homoserine lactones reveal unique structural features that dictate their ability to activate or inhibit quorum sensing. ChemBioChem. 2008;9:389–400. doi: 10.1002/cbic.200700551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mattmann ME, Shipway PM, Heth NJ, Blackwell HE. Potent and selective synthetic modulators of a quorum sensing repressor in Pseudomonas aeruginosa identified from second-generation libraries of N-acylated L-homoserine lactones. ChemBioChem. 2011;12:942–949. doi: 10.1002/cbic.201000708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eibergen NR, Moore JD, Mattmann ME, Blackwell HE. Potent and selective modulation of the RhlR quorum sensing receptor by using non-native ligands: an emerging target for virulence control in Pseudomonas aeruginosa. ChemBioChem. 2015;16:2348–2356. doi: 10.1002/cbic.201500357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bottomley MJ, Muraglia E, Bazzo R, Carfi A. Molecular insights into quorum sensing in the human pathogen Pseudomonas aeruginosa from the structure of the virulence regulator LasR bound to its autoinducer. J. Biol. Chem. 2007;282:13592–13600. doi: 10.1074/jbc.M700556200. [DOI] [PubMed] [Google Scholar]
- 31.Lintz MJ, Oinuma K-I, Wysoczynski CL, Greenberg EP, Churchill MEA. Crystal structure of QscR, a Pseudomonas aeruginosa quorum sensing signal receptor. Proc. Natl. Acad. Sci. U. S. A. 2011;108:15763–15768. doi: 10.1073/pnas.1112398108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang R-G, Pappas KM, Pappas T, Brace JL, Miller PC, Oulmassov T, Molyneaux JM, Anderson JC, Bashkin JK, Winans SC, Joachimiak A. Structure of a bacterial quorum-sensing transcription factor complexed with pheromone and DNA. Nature. 2002;417:971–974. doi: 10.1038/nature00833. [DOI] [PubMed] [Google Scholar]
- 33.Mattmann ME, Geske GD, Worzalla GA, Chandler JR, Sappington KJ, Greenberg EP, Blackwell HE. Synthetic ligands that activate and inhibit a quorum sensing regulator in Pseudomonas aeruginosa. Bioorg. Med. Chem. Lett. 2008;18:3072–3075. doi: 10.1016/j.bmcl.2007.11.095. [DOI] [PubMed] [Google Scholar]
- 34.Smith K, Bu Y, Suga H. Library screening for synthetic agonists and antagonists of a Pseudomonas aeruginosa autoinducer. Chem. Biol. 2003;10:563–571. doi: 10.1016/s1074-5521(03)00107-8. [DOI] [PubMed] [Google Scholar]
- 35.Zhu J, Beaber JW, Moré MI, Fuqua C, Eberhard A, Winans SC. Analogs of the autoinducer 3-oxooctanoyl-homoserine lactone strongly inhibit activity of the TraR protein of Agrobacterium tumefaciens. J. Bacteriol. 1998;180:5398–5405. doi: 10.1128/jb.180.20.5398-5405.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chai Y, Winans SC. Site-directed mutagenesis of a LuxR-type quorum-sensing transcription factor: alteration of autoinducer specificity. Mol. Microbiol. 2004;51:765–776. doi: 10.1046/j.1365-2958.2003.03857.x. [DOI] [PubMed] [Google Scholar]
- 37.Janssens JCA, Metzger K, Daniels R, Ptacek D, Verhoeven T, Habel LW, Vanderleyden J, De Vos DE, De Keersmaecker SCJ. Synthesis of N-acyl homoserine lactone analogues reveals strong activators of SdiA, the Salmonella enterica serovar Typhimurium LuxR homologue. Appl. Environ. Microbiol. 2007;73:535–544. doi: 10.1128/AEM.01451-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee J, Jayaraman A, Wood TK. Indole is an inter-species biofilm signal mediated by SdiA. BMC Microbiol. 2007;7:42–15. doi: 10.1186/1471-2180-7-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Michael B, Smith JN, Swift S, Heffron F, Ahmer BM. SdiA of Salmonella enterica is a LuxR homolog that detects mixed microbial communities. J. Bacteriol. 2001;183:5733–5742. doi: 10.1128/JB.183.19.5733-5742.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nguyen Y, Nguyen NX, Rogers JL, Liao J, MacMillan JB, Jiang Y, Sperandio V. Structural and mechanistic roles of novel chemical ligands on the SdiA quorum-sensing transcription regulator. mBio. 2015;6:e02429-02414-02410. doi: 10.1128/mBio.02429-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, Scheld M, Spellberg B, Bartlett J. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin. Infect. Dis. 2009;48:1–12. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.