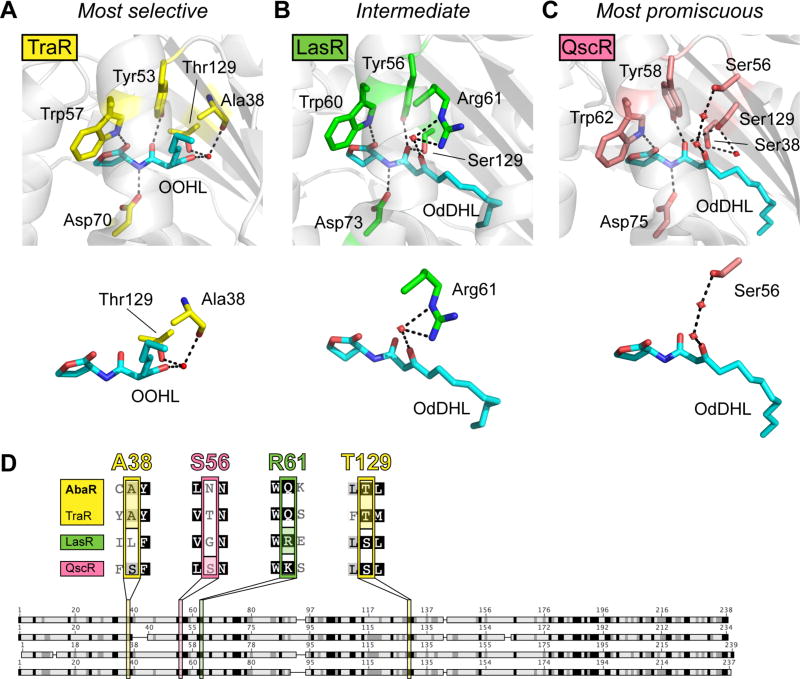
Fig. 3.
Model to explain AHL-selectivity in AbaR and LasR. (A) Ligand-binding pocket of X-ray crystal structure of TraR-OOHL (pdb 1L3L).32 (B) Ligand-binding pocket of X-ray crystal structure of LasR-OdDHL (pdb 2UV0).30 (C) Ligand-binding pocket of X-ray crystal structure of QscR-OdDHL (pdb 3SZT).31 Residues that form hydrogen bonds with the ligand are displayed on top, and simplified versions of binding pocket images displaying only the residues involved in hydrogen-bonding with the 3-oxo ligand moiety are shown below. Black dashed lines indicate hydrogen bonds, and red spheres indicate water molecules. (D) Alignment of AbaR with three 3-oxo-AHL-binding LuxR homologs. See Table S4 for full alignment. Darker shading indicates higher degree of conservation. Four residues are highlighted that bind the 3-oxo moiety of the proteins’ native AHL in reported X-ray crystal structures (A38 and T129 for TraR, S56 for QscR, and R61 for LasR).