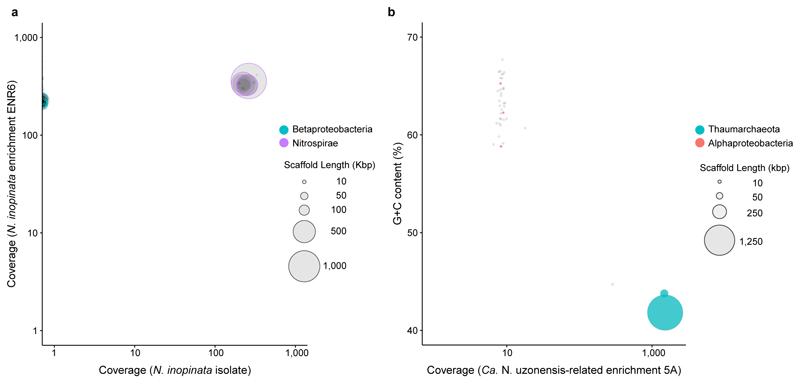
Extended Data Figure 1. Binning of the metagenome scaffolds from nitrifier cultures.
Circles represent scaffolds, scaled by the square root of their length. Only scaffolds ≥5 kbp are shown. Clusters of similarly coloured circles represent potential genome bins. a, Sequence composition-independent binning of the scaffolds from the N. inopinata isolate. The previous enrichment culture ENR6, which contained N. inopinata and only one contaminating betaproteobacterial organism1, was used for comparison in differential coverage binning. The lack of scaffolds from the betaproteobacterium (coverage = 0) shows the absence of this organism in the pure culture of N. inopinata. To further confirm that no trace amounts of the betaproteobacterium were left in the pure culture, the trimmed sequence reads were also mapped to the ENR6 metagenomic assembly that contained the contaminant1. None of 5.8 million reads mapped to the betaproteobacterium, whereas 99.8% of reads mapped to the closed genome of N. inopinata. Purity of the N. inopinata culture was also confirmed by fluorescence in situ hybridization, quantitative PCR targeting the amoA gene of N. inopinata and the soxB gene of the betaproteobacterium1, and the absence of growth in an organic medium. b, Binning of the scaffolds from the Ca. N. uzonensis-related enrichment culture 5A based on coverage and the G+C content of DNA. Aside from the Ca. N. uzonensis-related AOA strain 5A (99.93% 16S rRNA sequence identity), the culture contained an alphaproteobacterium at low abundance. Since the genome coverage of strain 5A was 1,512× and that of the alphaproteobacterium was 8×, the relative abundance of strain 5A in the enrichment culture was 99.5%.