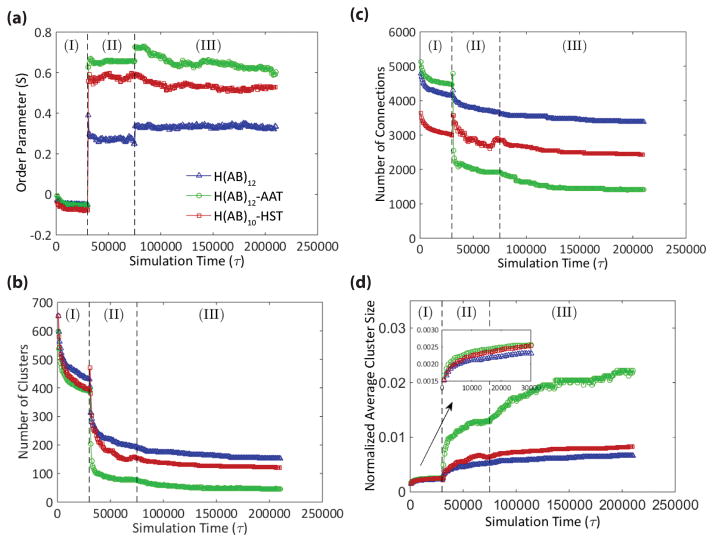
Figure 3. Quantification of the simulation results.
Each simulation consists of three stages: (I) initial equilibration, (II) shearing, and (III) post-shear equilibration. (a) Tracking the order parameter through the simulation shows that, in all cases, shearing results in greater alignment, or molecular order, in the direction of shear with H(AB)12-AAT aligning to the greatest degree followed by H(AB)10-HST, and lastly H(AB)12. (b, c) Conversely, there is a universal decrease in the number of clusters and connections after shearing with the rank order of the three peptides reversed from that of the order parameter. (d) The decreases in cluster and connection numbers are offset by cluster consolidation, which again follows the order observed for the order parameter. This suggests that molecular order is a means by which semicrystalline domains, modeled by the clusters, are consolidated into larger structures.