Figure 4. Novel small molecule ligand discovery against the I4/I6 binding site using ensemble docking.
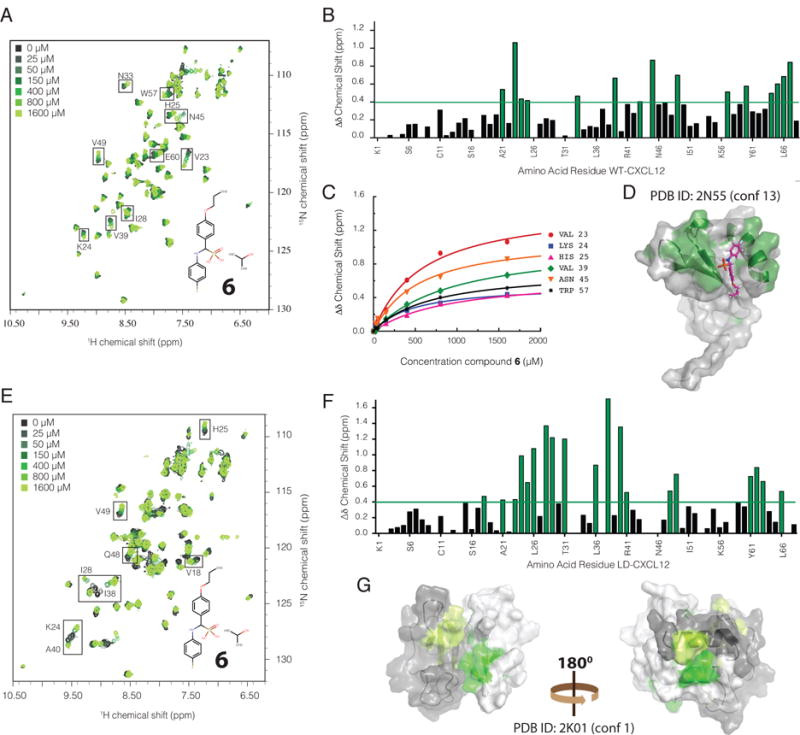
A. 1H-15N HMQC overlay of titration of WT-CXCL12 with 6 (inset) used for chemical shift mapping showing significant chemical shift perturbations for a subset of residues. B. Quantification of the change in chemical shift (Δδ Chemical Shift) for individual amino acid residues of CXCL12 with those experiencing the largest shifts highlighted in green. C. Fitted curves for selected residues used to calculate the affinity (Kd) of 6 for WT-CXCL12. D. Mapping of significantly perturbed residues onto the NMR structure (conformation 13) of constitutively monomeric CXCL12 (L55C/I58C) (PDB ID: 2N55) used to identify 6 in ensemble virtual screening, corresponds to the predicted binding site for 6. E. 1H-15N HMQC overlay of titration of locked dimeric CXCL12 (L36C/A65C) with 6 (inset) used for chemical shift mapping showing significant chemical shift perturbations for a subset of residues. F. Quantification of the change in total chemical shift (1H/15N Δδ Chemical Shift) for individual amino acid residues of LD-CXCL12 (locked dimer) with those experiencing the largest shifts highlighted in green. G. Mapping of significantly perturbed residues onto the NMR structure (conformation 1) of dimeric CXCL12 (L36C/I65C) (PDB ID: 2K01). The two monomers are colored in gray and white with corresponding perturbed residues colored in light green and dark green respectively.
