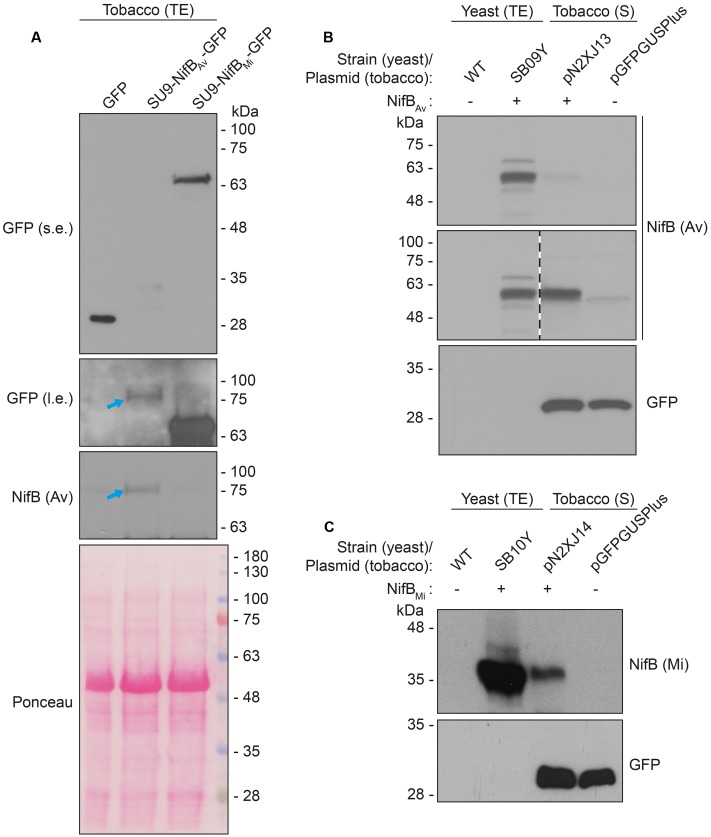
FIGURE 6.
Expression and solubility of mitochondria targeted (SU9) NifBAv and NifBMi in N. benthamiana leaves. (A) Western blot analysis of total protein extracts (TE) prepared from infiltrated N. benthamiana leaves expressing GFP, SU9-NifBAv-GFP or SU9-NifBMi-GFP. Blue arrows indicate the polypeptide recognized both by GFP and NifBAv specific antibodies. Short (s.e.) and long (l.e.) film exposures of the GFP antibody probed membrane are shown. (B) Migration of SU9-NifBAv-His10 when expressed in S. cerevisiae and N. benthamiana. Migration in SDS-PAGE was determined after Western blot analysis using NifBAv specific antibodies. Total protein extracts (TE) from W303-1a S. cerevisiae cells (WT) or cells expressing SU9-NifBAv-His10 (SB09Y) were prepared. Soluble protein extracts (S) from N. benthamiana leaf cells infiltrated with A. tumefaciens containing control vector (pGFPGUSPlus) or vector for expression of SU9-NifBAv-His10 (pN2XJ13). Dotted line indicate different exposures of the right part of the membrane. See Supplementary Figure S9 for entire gel of the cropped exposure. (C) Migration of SU9-NifBMi-His10 when expressed in S. cerevisiae and N. benthamiana. Migration in SDS-PAGE was determined after Western blot analysis using NifBMi specific antibodies. Total protein extracts (TE) from W303-1a S. cerevisiae cells (WT) or cells expressing SU9-NifBMi-His10 (SB10Y) were prepared. Soluble protein extracts (S) from N. benthamiana leaf cells infiltrated with A. tumefaciens containing control vector (pGFPGUSPlus) or vector for expression of SU9-NifBMi-His10 (pN2XJ14). As control of N. benthamiana leaf infiltration, GFP expressed from the pGFPGUSPlus vector backbone was detected (B,C).