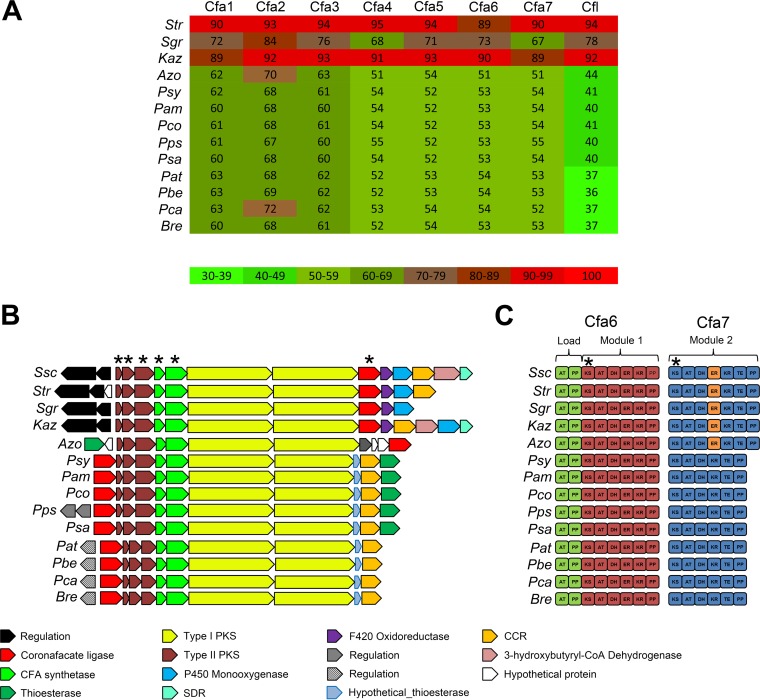
FIG 4.
(A) Heat map showing the protein BLAST identity hits to the S. scabiei 87-22 Cfa1-7 and Cfl proteins in different bacterial genomes. The actual percent amino acid identity is shown in each square. Abbreviations are as follows: Str, Streptomyces sp. NRRL WC-3618; Sgr, Streptomyces griseoruber DSM40281; Kaz, Kitasatospora azatica ATCC 9699; Azo, Azospirillum sp. B510, Psy, Pseudomonas syringae pv. tomato DC3000; Pam, Pseudomonas amygdali pv. morsprunorum, Pco, Pseudomonas coronafaciens pv. porri LMG 28495; Pps, Pseudomonas psychrotolerans NS274; Psa, Pseudomonas savastanoi pv. glycinea B076; Pat, Pectobacterium atrosepticum SCRI1043; Pbe, Pectobacterium betavasculorum NCPPB 2795; Pca, Pectobacterium carotovorum subsp. carotovorum UGC32; Bre, Brenneria sp. EniD312. (B) Organization of known and putative CFA biosynthetic gene clusters identified in sequenced bacterial genomes. Related genes are in the same color, and the known or predicted functions are indicated below. The genes that were used for construction of the concatenated phylogenetic tree (Fig. 6A) are denoted by an asterisk. Ssc, Streptomyces scabiei 87-22. (C) Domain organization of the Cfa6 and Cfa7 type I polyketide synthases encoded in the CFA biosynthetic gene clusters from different bacteria. The different modules (load, modules 1 and 2) are indicated as well as the enoyl reductase (ER) domain that is found only in a subset of Cfa7 proteins (indicated in orange). The domain nucleotide sequences that were used for construction of the concatenated phylogenetic tree (Fig. 6A) are denoted by an asterisk. Abbreviations are as follows: AT, acyltransferase; PP, phosphopantheinate; KS, ketosynthase; DH, dehydratase; KR, ketoreductase; TE, thioesterase.