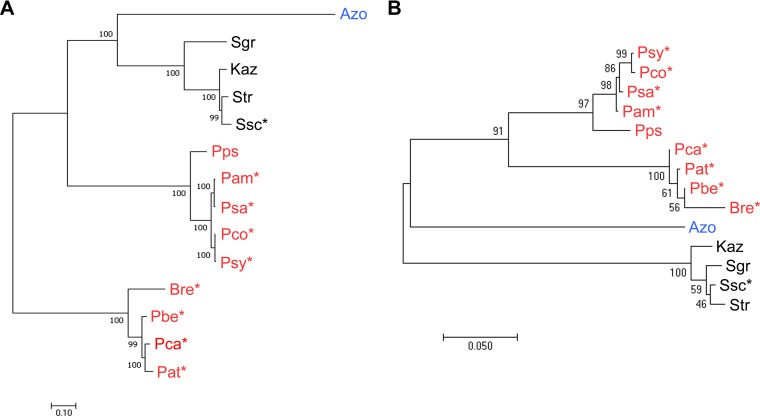
FIG 6.
Maximum likelihood phylogeny of the core CFA biosynthetic genes (A) and the 16S rRNA gene (B) from different bacteria. The CFA biosynthetic gene cluster tree was constructed using the concatenated cfa1–5, cfl, cfa6 (KS domain), and cfa7 (KS domain) nucleotide sequences. Bootstrap values of ≥50% are shown at the respective branch points and are based on 100 repetitions for the CFA gene cluster tree and 1,000 repetitions for the 16S rRNA gene tree. The scale bars indicate the number of nucleotide substitutions per site. Gammaproteobacteria are in red, Alphaproteobacteria are in blue, and Actinobacteria are in black. Known plant-pathogenic organisms are indicated with an asterisk (*). Abbreviations are as described for Fig. 4.