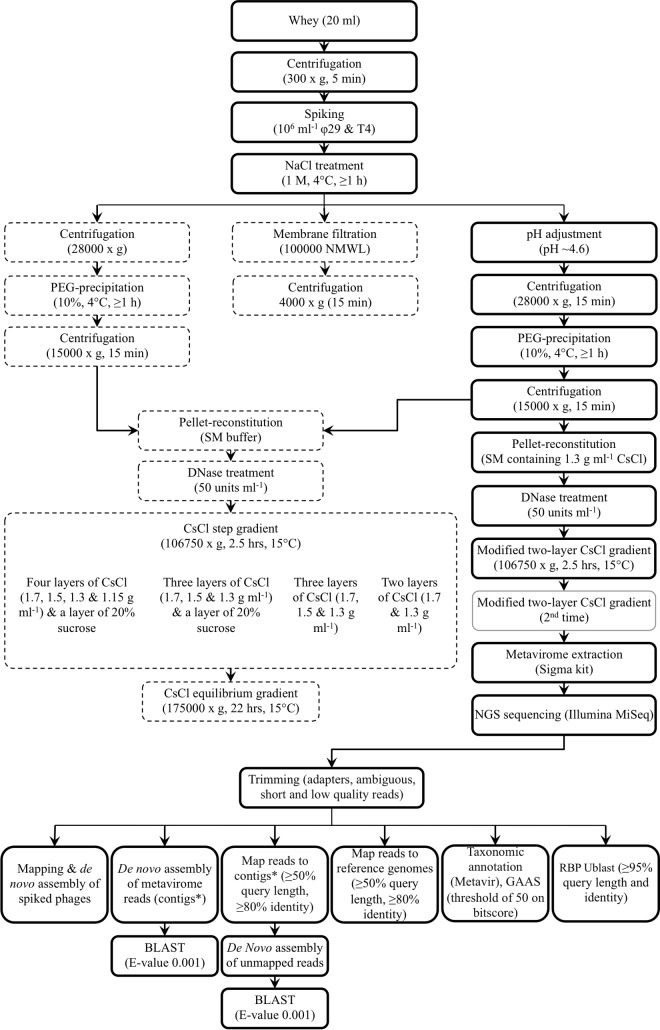
FIG 1.
Dairy phage metagenomic analysis workflow: overview of method optimization and analysis. Different techniques were assessed in terms of ability to separate bulk proteins and to recover spiked Bacillus subtilis φ29 and Escherichia coli T4 phages from whey samples. Broken-line boxes represent pathways with inadequate protein removal or heavy loss of φ29 and T4. Continuous boldface line boxes represent pathways with removal of a reasonable amount of proteins and/or recovery of a majority of the spiked phages. A continuous faded-line box represents an optional step that depends on the outcome of the previous step. The modified two-layer CsCl gradient was made of a layer of 1.7 g ml−1 CsCl in SM buffer overlaid by a layer of phage suspension (PEG pellet in 1.3 g ml−1 CsCl solution). The method's overall recovery efficiency was assessed following a CsCl equilibrium gradient or the last modified CsCl gradient ultracentrifugation. Local databases containing the genomes of dairy Lc. lactis and Leuconostoc phages and phylogenetically organized receptor-binding protein (RBP) groups were used to perform the respective mapping and Ublast analyses. NMWL, nominal molecular weight limit; PEG, polyethylene glycol; SM, sodium-magnesium (buffer); NGS, next-generation sequencing; GAAS, genome relative abundance and average size.