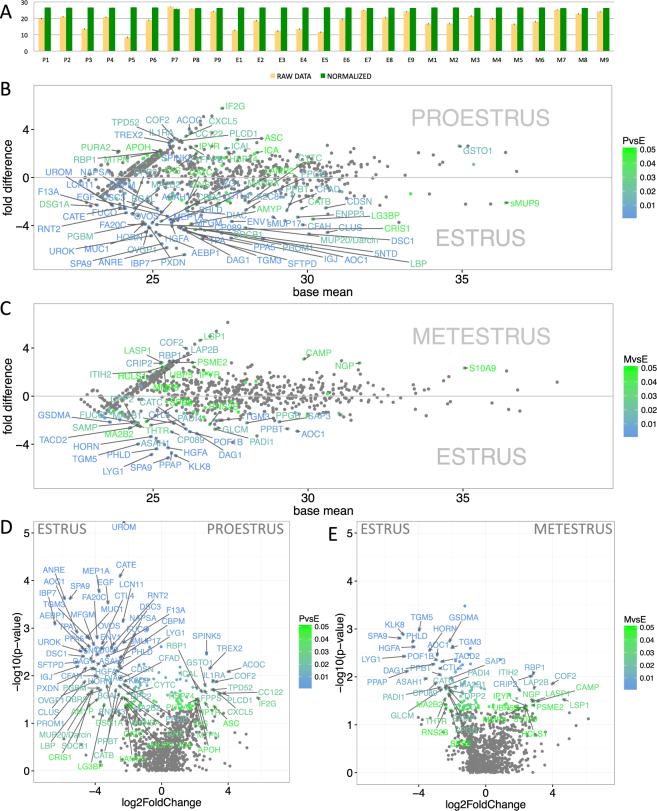
Figure 1.
Graphical representation of differentially expressed/abundant proteins. Before normalization, the data revealed some variation between individuals (A–yellow bars). However, after the quantile-normalization procedure (A–green bars), the mean value and standard error bars show almost no variation between the samples. Significant differentially-expressed proteins are demonstrated with MA plots and are more common in estrus than in proestrus or metestrus (B,C). PLGEM model was involved in testing the differences in normalized signal values between proestrus and estrus (B), and between metestrus and estrus (C). The level of significance (PvsE, MvsE) is scaled from green (P < 0.05) to blue (P < 0.01) and only the data points with FC > 2 are annotated. The x-axis represent the basal mean of signal intensities in B and C. The dependence of particular fold changes on p-values is provided using the volcano plots in D and E.