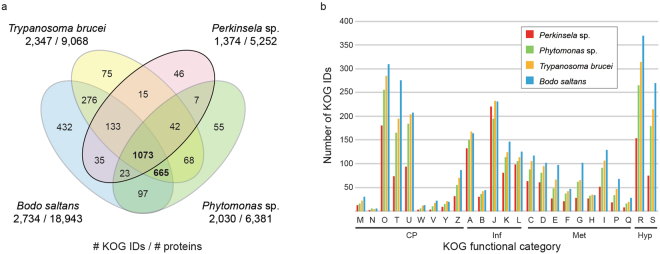
Figure 2.
Functional diversity of proteins in Perkinsela sp. compared to other kinetoplastids. (a) Venn diagram shows overlap in the number of proteins assigned a KOG ID encoded in the nuclear genome of Perkinsela sp. CCMP1560/4, the free-living flagellate Bodo saltans 25, the human pathogen Trypanosoma brucei 27, and the plant pathogen Phytomonas sp.26. Functions were assigned based on the euKaryotic Orthologous Groups (KOG) database29. The total number of proteins predicted from the nuclear genome of each organism is also shown. (b) Histogram showing the unique numbers of KOG IDs found in Perkinsela sp., Phytomonas sp., T. brucei and B. saltans. KOG categories are as follows: A, RNA processing and modification; B, chromatin structure and dynamics; C, energy production and conversion; D, cell cycle control, cell division and chromosome partitioning; E, amino acid transport and metabolism; F, nucleotide transport and metabolism; G, carbohydrate transport and metabolism; H, coenzyme transport and metabolism; I, lipid transport and metabolism; J, translation, ribosomal structure and biogenesis; K, transcription; L, replication, recombination and repair; M, cell wall, membrane or envelope biogenesis; N, cell motility; O, post-translational modification, protein turnover, chaperones; P, inorganic ion transport and metabolism; Q, secondary metabolites biosynthesis, transport and catabolism; R, general function prediction only; S, function unknown; T, signal transduction; U, intracellular trafficking, secretion and vesicular transport; V, defence mechanisms; W, extracellular structures; Y, nuclear structure; Z, cytoskeleton. Higher KOG categories are as follows: CP, cellular processing and signalling; Hyp, poorly characterized; Inf, information storage and processing; Met, metabolism.