Figure 3. Genetic or pharmacologic disruption of the transcriptional regulatory apparatus down-regulates Mcl-1 in bortezomib-sensitive or -resistant MM cells.
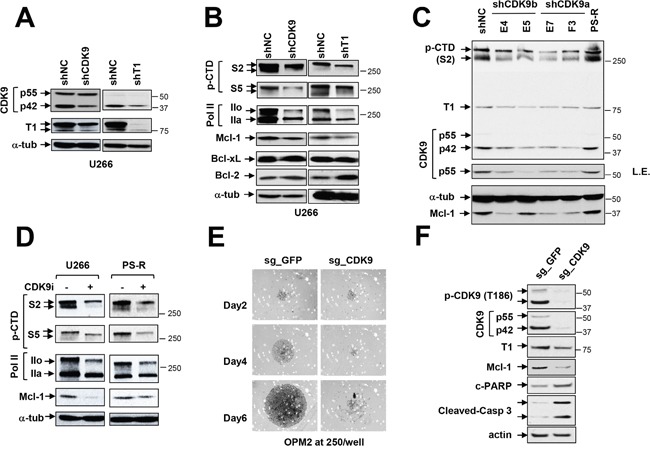
(A-B) U266 cells were stably transfected with constructs encoding shRNA targeting CDK9 (shCDK9) or cyclin T1 (shT1) or scrambled sequence as a negative control (shNC). Immunoblotting analysis was performed to profile basal levels of CDK9, cyclin T1, phosphorylated forms (serine-2 and 5, CTD) of RNA pol II, Mcl-1, Bcl-2, and Bcl-xL in shCDK9 and shT1 cells as described previously. α-tubulin controls were assayed to ensure equivalent loading and transfer. Replicate experiments yielded equivalent results. (C) PS-R (bortezomib-resistant U266) cells were stably transfected with constructs encoding shRNA targeting CDK9 (shCDK9a or shCDK9b), and subjected to immunoblotting analysis to monitor basal levels of CDK9, cyclin T1, phosphorylated (serine-2, CTD) RNA pol II and Mcl-1. Lanes were loaded with 30 μg of protein; α-tubulin controls were assayed to ensure equivalent loading and transfer. Duplicate experiments yielded equivalent results. L.E. indicates long exposure. (D) U266 and PS-R cells were treated with CDK9i 15 μM for 24 hr, after which expression of phosphorylated (serine-2 and 5, CTD) RNA pol II, pol II, and Mcl-1 was monitored by immunoblotting analysis as in C. α-tubulin controls were assayed to ensure equivalent loading and transfer. Duplicate experiments yielded equivalent results. (E) OPM2 MM cells were infected with lentivirus encoding Cas9 and sgRNA targeting GFP or CDK9. After infection and selection with puromycin (1.5 mg/ml, 48 hr), cells were seeded in a 48-well round-bottom plate (250 cells per well), and images were obtained on day 2, 4 and 6. Images were obtained with an IX71-Olympus research inverted system microscope at 40× magnification. (F) Protein extracts were obtained from non-targeting and sgCDK9 cells, and immunoblotting analysis performed to monitor expression of CDK9 (55 and 42 kDa), cyclin T1, Mcl-1, and cleaved PARP and caspase 3. Lanes were loaded with 30 μg of protein; β-actin controls were assayed to ensure equivalent loading and transfer. Duplicate experiments yielded equivalent results.
