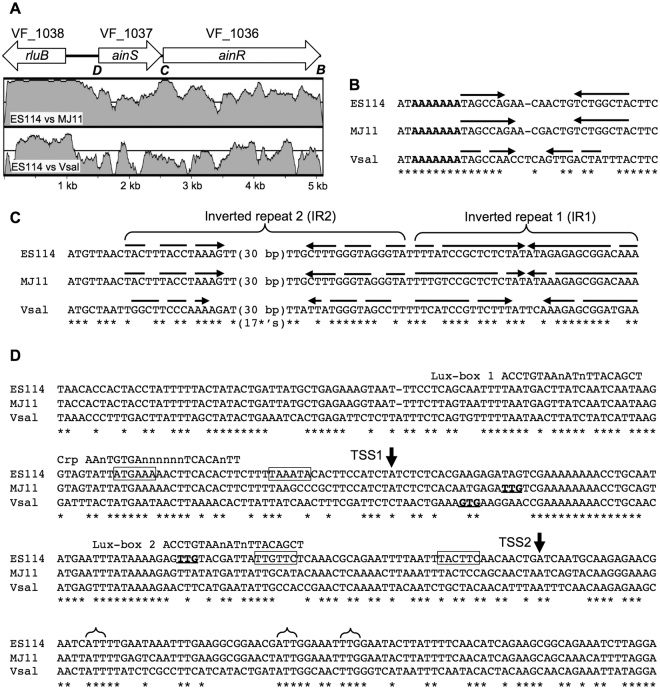
Figure 2.
Comparison of the ainSR locus in three V. fischeri and V. salmonicida strains. The sequence between the stop codons of VF_1038 and VF_1035 in V. fischeri ES114 was compared to the orthologous regions from V. fischeri MJ11 and V. salmonicida LFI1238. In panel A, arrows show the arrangement of the three complete genes at this locus, which extends to the stop codon of VF_1035 on the right. Bold and italicized letters under the arrows indicate regions for which DNA sequence is shown in the corresponding panels. Homology between ES114 and MJ11 or V. salmonicida is shown for corresponding regions in shaded plots that range from fifty to one hundred percent identity within a 100-bp window as determined by VISTA73 with the LAGAN74 alignment function and default settings. The grey line denoted seventy-five percent identity. Panel B shows a conserved putative Rho-independent transcriptional terminator downstream of VF_1035 that was identified by ARNold42, with arrows indicating inverted repeat stems, with stem mismatches indicated by gaps on the arrows. A bold-lettered run of A’s indicates the canonical string of U’s (on the reverse strand transcript) following a stem loop structure in such terminators. Panel C shows sequences aligned from the start codon of ainR, with arrows indicating inverted repeats and mismatches indicated by gaps on the arrows. Panel D shows an alignment of sequences upstream of and within ainS. Two transcriptional start sites mapped in strain ES114 by 5′ RACE are indicated as TSS1 and TSS2, and putative −10 and −35 promoter elements associated with TSS1 and TSS2 are boxed. Start codons predicted by The SEED44 are indicated with bold and underlined letters. Brackets indicate alternative start codons conserved across all three strains. A CRP binding site24 and possible “lux box” LuxR-binding sequences are highlighted by alignment with the respective consensus binding sequence. In panels B–D, asterisks indicate bases conserved in all three strains.