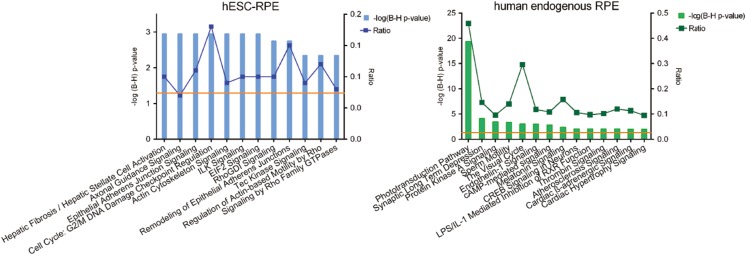
Fig. 6.
Canonical pathways identified by IPA for the genes that are significantly differentially expressed between the hESC-RPE cells and the human endogenous RPE. The left graph (blue) depicts the canonical pathways that relate to the genes specifically expressed in the hESC-RPE (this study). The right graph (green) depicts the canonical pathways that relate to the genes specifically expressed in the human endogenous RPE (submitted). In the graphs, the left y-axis displays the –log of the Benjamini-Hochberg corrected –value. The right axis displays the ratio of the number of genes derived from our dataset, divided by the total number of genes in the pathway. The bars show the –log(B-H) p-value. The orange line indicates the threshold at a B-H corrected p-value <0.05