Fig. 2.
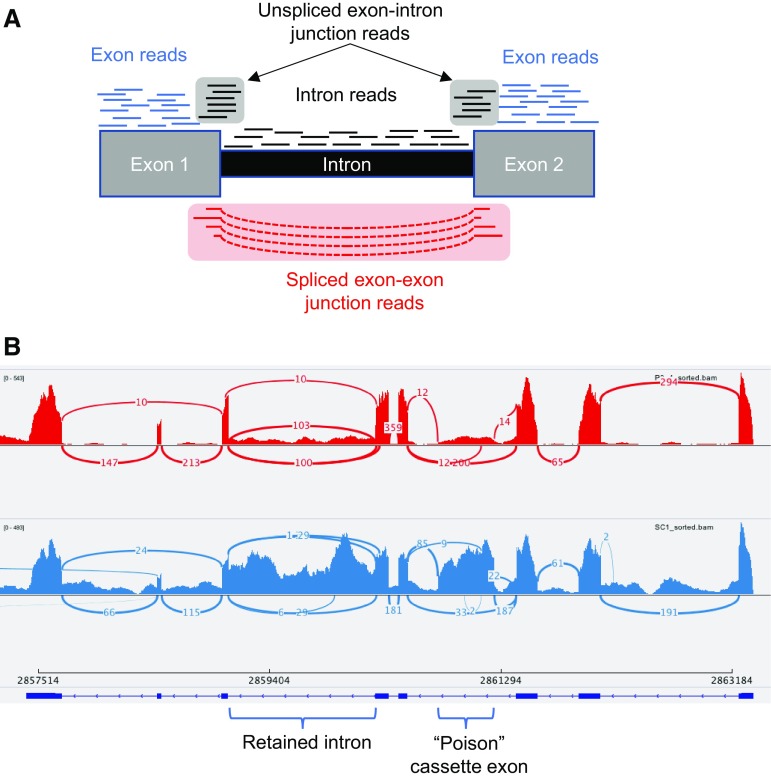
Intron retention profiling by mRNA-Seq. a Schematic diagram showing distribution of sequence reads informative for intron retention. Percent intron retention can be calculated from the ratio of unspliced exon–intron junction reads to total junction reads (unspliced exon–intron and spliced exon–exon), or from the read density across the intron compared to adjacent exons. Uniform read density across the intron rules out alternative processing events. b Example of mRNA-Seq data from rat primary aorta smooth muscle cells (unpublished data). Differentiated, blue lower panel; proliferative, red upper panel. The Sashimi plot, generated from the Integrative Genomics Viewer (Robinson et al. 2011), shows the Srsf7 gene. In differentiated cells, there is substantial IR in intron 6, as well as inclusion of the known “poison” cassette exon between protein coding exons 3 and 4 (Lareau et al. 2007)