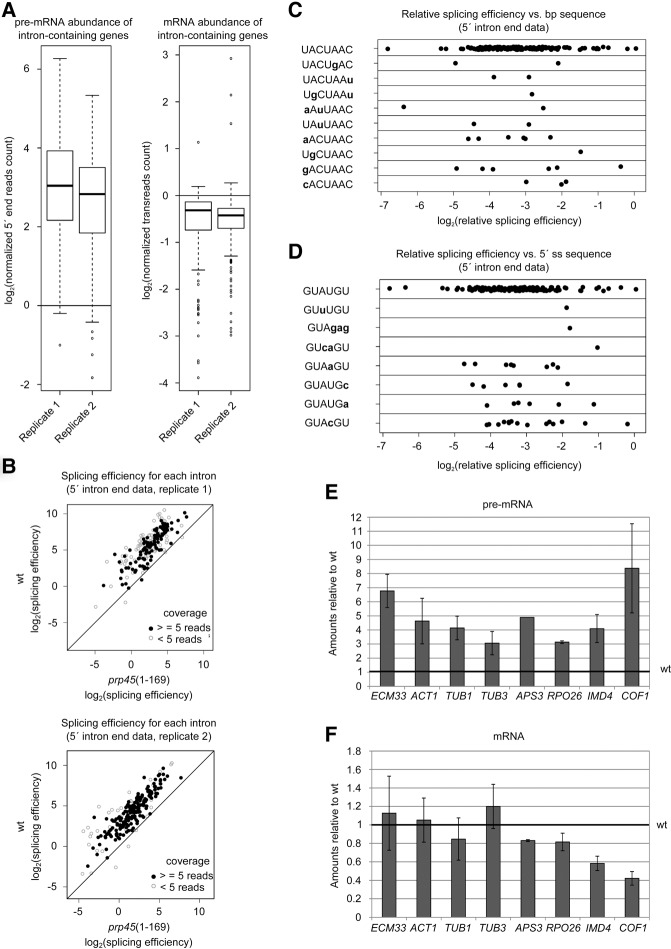
FIGURE 1.
Impairment of Prp45 function leads to pre-mRNA accumulation. (A–C) Transcriptome analysis of prp45(1–169) cells in comparison with WT in two biological replicates. (A) pre-mRNA is accumulated in the prp45 mutant (left panel), while the mRNA levels of intron-containing genes are affected only to a limited extent (right panel). The bottom and top of the box represent the second and third quartiles, respectively; the line represents the median. Whiskers extend up to 1.5× of the interquartile range. (B) The overall relative splicing efficiency is impaired in prp45(1–169) cells. However, this drop in relative splicing efficiency does not show any tendency with respect to the branch point (C) or 5′ss sequence (D). Graphs C and D show results from one biological replicate. Ratios of mutant/WT splicing efficiencies are plotted. (E,F) RT-qPCR quantification of splicing of selected genes. Spliced (E) and unspliced (F) transcripts were quantified by RT-qPCR and normalized to TOM22. Data are expressed relative to WT levels. Error bars represent standard deviations (SD) of at least three biological experiments.