FIGURE 5.
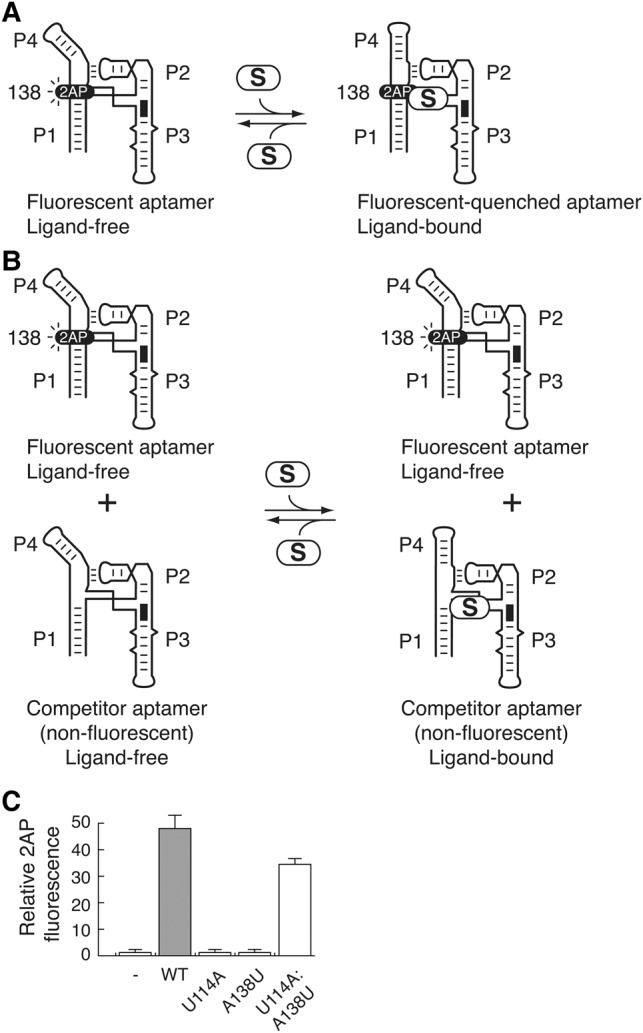
U114A and A138U mutants do not efficiently perform SAM binding. (A) Schematic representing the fluorescence emission of a 2-aminopurine (2AP) fluorophore at position 138 of the SAM aptamer. A wild-type aptamer containing a 2AP at position 138 is used as a fluorescent reporter that is quenched upon SAM binding. In the presence of Mg2+, the aptamer is folded in a conformation allowing 2AP fluorescence. However, SAM (S) binding to the aptamer results in the P1–P4 helical stacking and 2AP fluorescence quenching. (B) Schematic representing the competition assay in which the fluorescent aptamer is incubated in the presence of a nonfluorescent competitor aptamer. The binding of SAM by the competitor allows the fluorescent aptamer to remain free from SAM binding, thus leading to a 2AP fluorescence increase. (C) Relative binding (%) is reported as a function of competitor aptamer. In the absence of any competitor (−), very little fluorescence can be detected due to the efficient 2AP fluorescence quenching that occurs through SAM binding. The relative binding is shown for the wild-type (WT), U114A, A138U, and U114A:A138U aptamer competitors.
