(
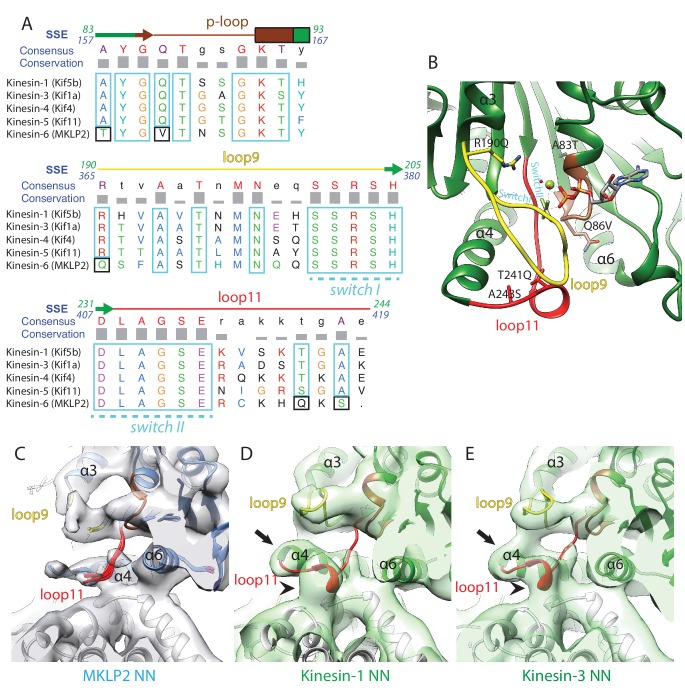
A) Sequence alignment of important regions at the nucleotide pocket for kinesin-6 MKLP2 and representative members from other Kin1/3/4/5 for which X-ray structures are available. Residue letters are coloured according to their properties according to the Clustal X scheme (
Larkin et al., 2007). Information on consensus, conservation and secondary structure is shown above the alignments. Sequence numberings for Kif5B (Kin1, green) and MKLP2 (kinesin-6, blue) are shown adjacent to the secondary structure schematics. Highly conserved/similar kinesin residues are boxed in light blue, whereas highly conserved kinesin residues which have diverged are boxed in black. The signature kinesin switch I and II sequences are indicated. (
B) The nucleotide pocket of the X-ray structure of tubulin-bound Kin1 in the presence of ADP.AlFx (PDB 4HNA, [
Gigant et al., 2013]). The position of the switch I/II sequences are annotated in cyan, whilst those that have diverged in MKLP2 are shown in sticks and annotated in black, in format
Kin1 amino-acid,
Kin1 amino-acid number, mouse MKLP2 amino-acid. (
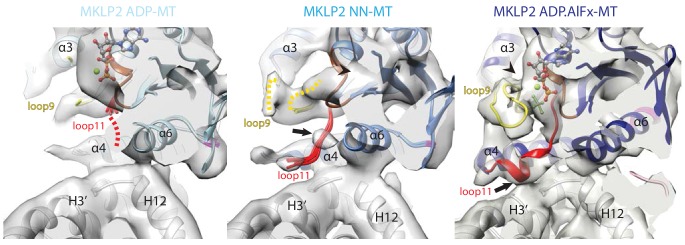
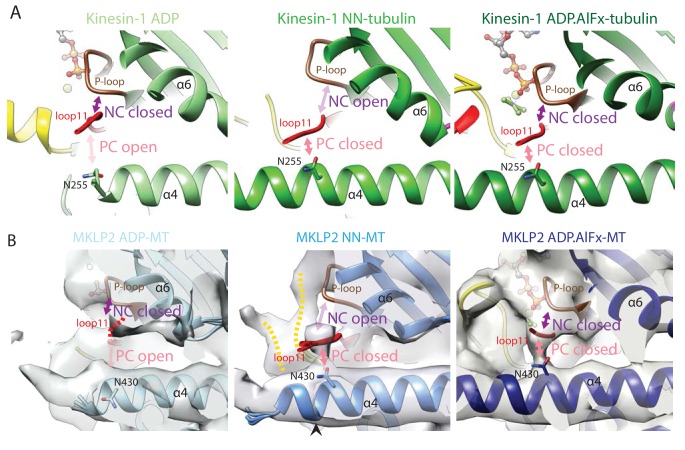
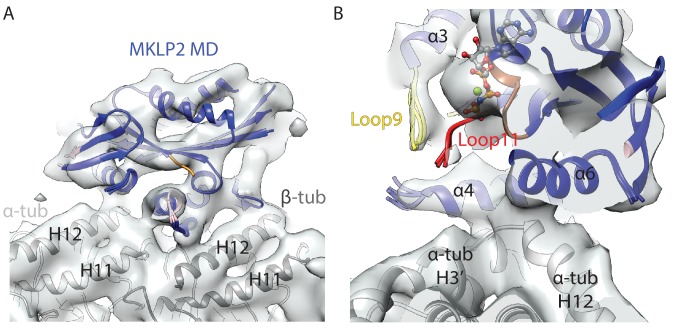
C–E) The nucleotide binding regions in the NN states of (
C) kinesin-6 MKLP2, (
D) Kin1 Kif5a and (
E) Kin3 Kif1A (
Atherton et al., 2014) are shown in their corresponding experimental densities (grey density for MKLP2-MD, green densities for Kin1 and Kin3). Arrows indicate full extension of helix-α4’s N-terminus in this state and arrowheads indicate formation of a single-turn helix in loop11 for Kin3 and Kin1 that contacts the MT, in contrast to MKLP2 where these regions are less ordered.